Capture Kits
The capture kit is a targeted sequencing panel used in targeted selection. It is designed for highly specific cleavage of the genomic regions of interest. The capture kits are used in the analysis of samples obtained from targeted sequencing. On "Capture kits" page, there are built-in and custom (if uploaded) capture kits. Both can be used for sample analysis. For targeted sequencing sample analysis, you can either select the corresponding capture kit when uploading the sample, or the capture kit that best suits the sample will be automatically selected from all capture kits presented in the system. In addition, the capture kit can be used to filter SNVs/Indels discovered in the sample.
To open the page with capture kits, go to "Capture kits" page from "Settings" page block through the navigation panel on the left:
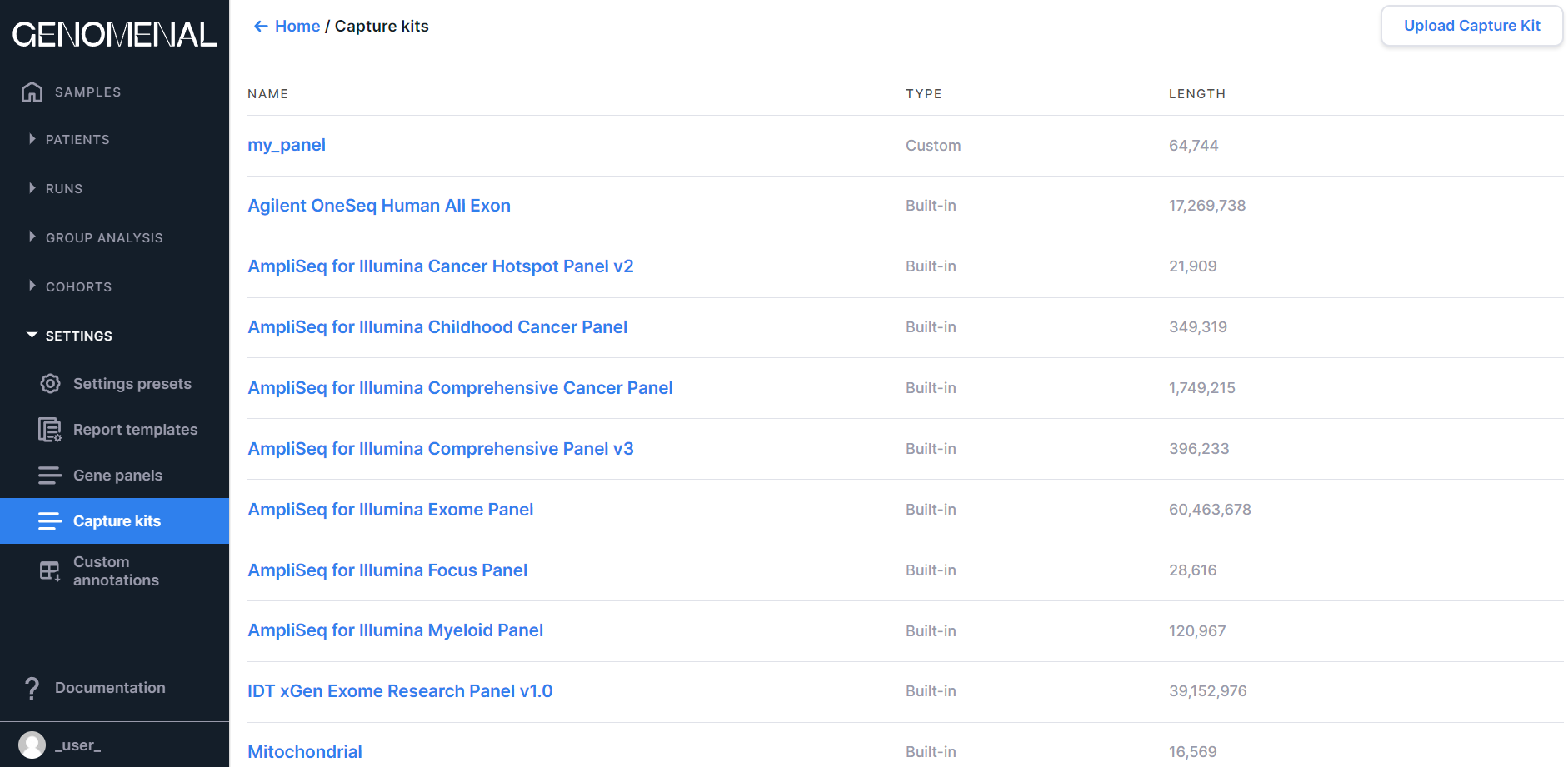
On the page, there is a table with the following columns:
- "NAME" is the capture kit name (the name of the uploaded capture kit file);
- "TYPE" is the capture kit type: Built-in (standard kits, often used in sequencing and built into the system) or Custom (uploaded by the user);
- "LENGTH" is the total coverage length of the capture kit (bp).
Upload Capture Kit#
- Click on
to open the capture kit upload window:

- By default, the reference genome assembly corresponding to the genomic intervals of the uploaded capture kit is detected automatically by Genomenal. You can determine the assembly by clicking on the appropriate field and selecting the assembly in the drop-down list: hg19 or hg38.
- Upload the capture kit file in BED format. The capture kit file is a tab-delimited or space-delimited file with three required columns: chromosome, start interval position on the chromosome, end interval position on the chromosome. To upload, drag and drop capture kit file into the drop-area or click on it to open the File Browser:

- Click on
 .
.
The uploaded capture kit will be displayed in the table with "Custom" type. If the reference capture kit genome assembly was detected as hg19, then for this capture kit, a liftover is performed, i.e., the interval genome positions are converted from hg19 to hg38. Capture kits that have been uploaded as hg19 are marked as "Mapped to hg38" in the table.
Download Capture Kit#
To download a capture kit file in BED format, click on capture kit name in "Name" column.
To download the uploaded capture kit file in BED format (the file before processing,
including liftover), hover over its row and
click on
(this option is available only for custom kits).
Delete Capture Kit#
You can delete only custom capture kit. To do this, hover over capture kit row and
click on .