Profile Settings
The "Profile settings" page contains your personal settings: account settings (email, login, password), the Program navigation panel settings, patient page settings, SNV Viewer and CNV Viewer settings.
To open "Profile settings" page, hover over your username at the bottom of the navigation panel
and click on the three
dots .
In the opened menu, select "Profile":
You will see a settings page:

Account settings#
Email address#
Здесь вы можете посмотреть, какой email вы использовали при регистрации в Системе. Для того чтобы его поменять, нажмите на , а затем в открывшемся окне введите новый адрес почты и ваш действующий пароль аккаунта для подтверждения действия:
Here you can see what email you used when registering in the System. In order to change it,
click on , and then in the
window that opens, enter a new email address and your current account password to confirm the action:

After filling out the form, click on .
Now check the inbox of the email that you indicated as new, and confirm the change of address in the letter.
Login#
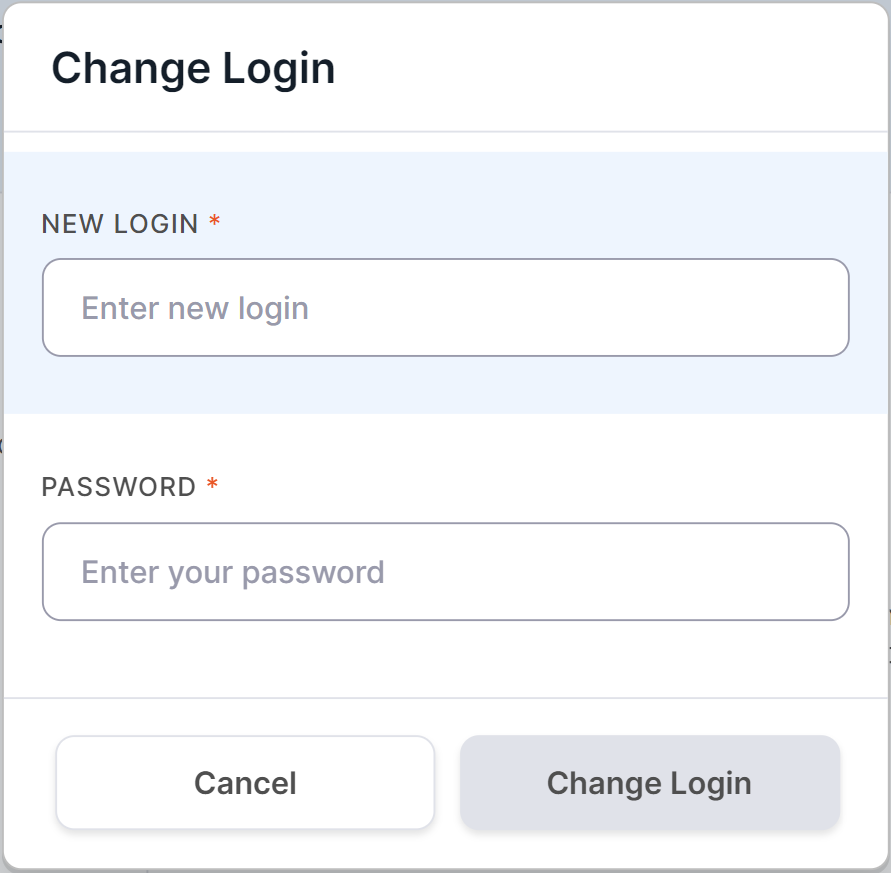
Here you can see which login you used when registering in the System. In order to change it,
click on , and then in the
window that opens, enter a new login and your current account password to confirm the action:
After filling out the form, click on  .
Login successfully changed!
.
Login successfully changed!
Login can consist of any characters up to 128.
Password#
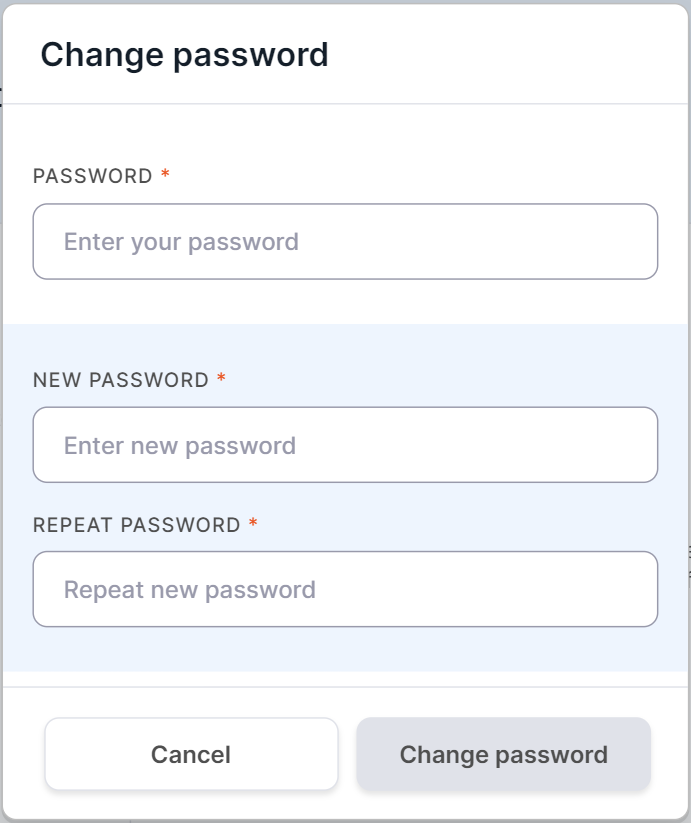
To change the password for logging into the System,
click on , and then in
the window that opens, enter your current account password to confirm the action and enter the new password twice:
After filling out the form, click on .
Password successfully changed!
Password can consist of any characters up to 128.
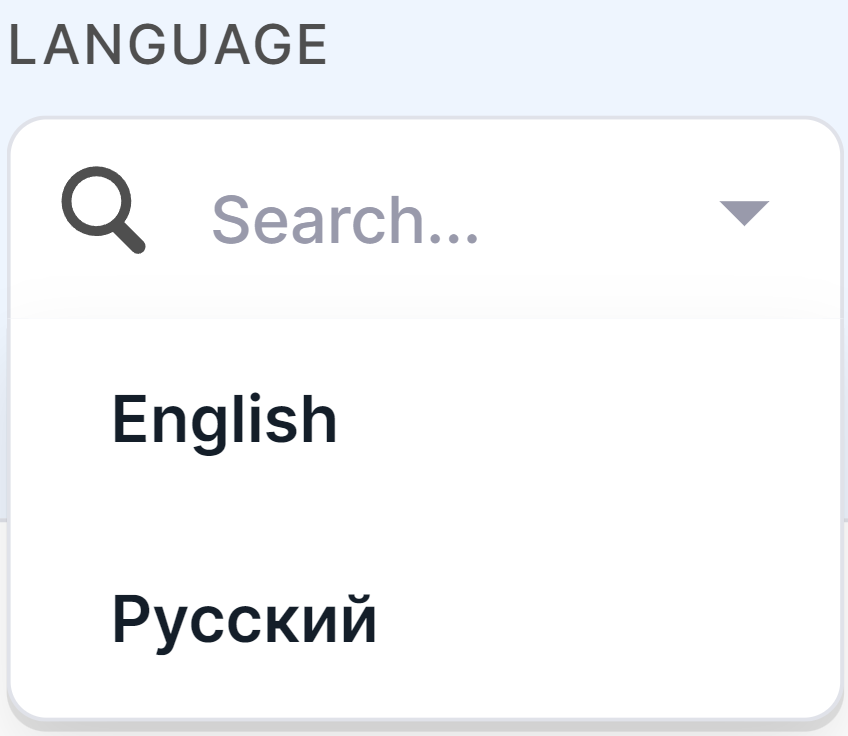
Program Language#
Here you can choose the Program language that is more convenient for you. To do this, click on the language selection field and select the language in the drop-down list:
Notification about processing completion#
Here you can configure receiving notifications when sample processing and group analyzes are complete. Sample analysis completion notifications are sent when all samples uploaded into a single patient or run have completed processing. When you restart processing a sample or group analysis, you will also receive a completion notification. Notifications are sent to the email chosen in your account settings. In the email, you will find a link to the page of the patient or run whose sample processing has completed, or a link to the completed group analysis. For patients and runs, links to samples whose processing completed successfully or with failures will be provided. To receive notifications when sample processing or group analyzes are completed, enable the corresponding option "Notify when processing is finished".
Navigation panel#
The section displays which page sections are shown on the Program navigation panel in addition to the home page "Samples" (all are selected by default):
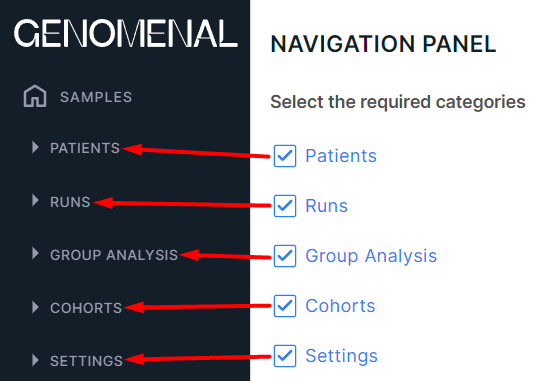
To remove a section from the panel, uncheck it.
Patient page#
There can be two tabs on the patient page: "Main" and "CNV Report" if "CNV Report" has been generated (see conditions here):
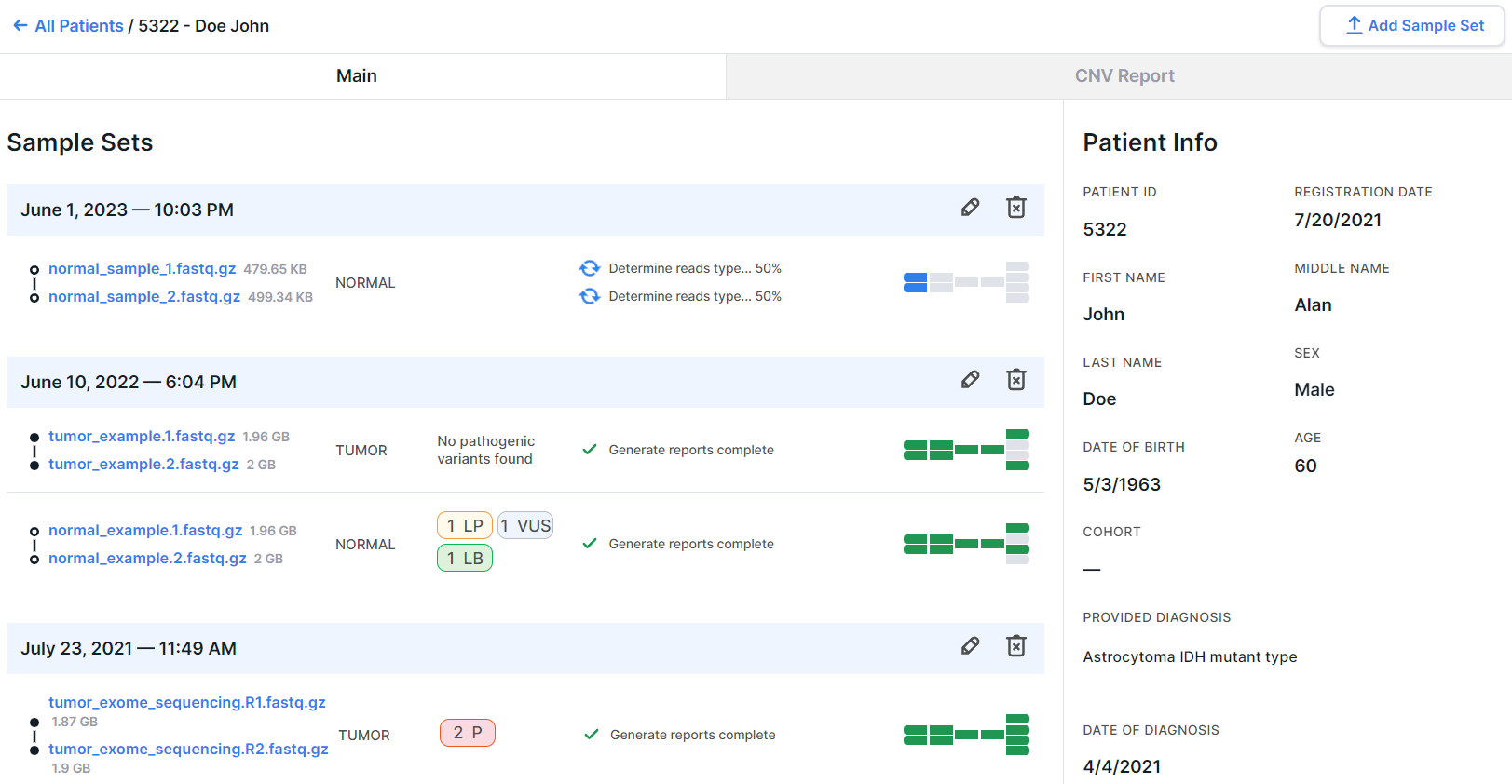
In this section, you can configure which tab you will see first when you open the patient page:
- the "Main" tab, which contains information about the patient and uploaded sample sets (chosen by default);
- the "CNV Report" tab with a report on the results of the copy number analysis in the patient samples.

info
If the patient "CNV Report" has not been generated at all or is still being generated, then even if you have selected the "CNV Report" tab here, the "Main" tab will open.
SNV Viewer#
SNV Viewer is an embedded service for viewing and analyzing variants. It is described in detail in the corresponding section.
Filtering mode#
SNV Viewer has two variant filtering modes: Basic with basic filters and Advanced with more filters and the ability to compose complex queries with multiple filter conditions.
In this section, you can configure which filtering mode you will see when you first open the sample SNV Viewer page (Basic Filtering chosen by default):

Filters in Basic Filtering mode#
The default filters are described here:

You can change the choice of filters (remove or add new ones):
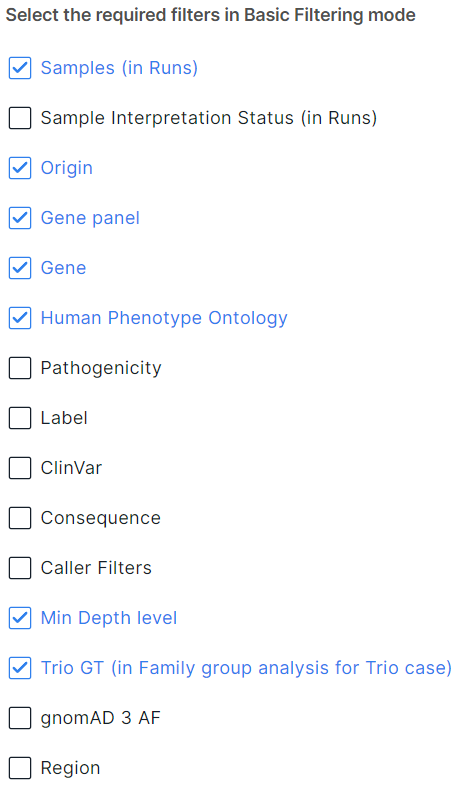
Available filters:
- Samples is a filter of variants by run samples in which these variants were discovered. Displayed only in the run SNV Viewer. Added by default.
- Sample Interpretation Status is a filter of variants by the interpretation status of the run sample in which these variants were discovered (completed or uncompleted interpretation). Displayed only in the run SNV Viewer.
- Origin is a variant filter by mutation type: Somatic or Germline. Added by default. Not displayed in SNV Viewer with germline mutations and the group analysis SNV Viewer.
- Gene panel is a variant filter by gene panel. Added by default.
- Gene is a variant filter by gene transcripts in which variants are located. Added by default.
- Human Phenotype Ontology (HPO) is a variant filter by genes associated with terms from a vocabulary of phenotypic abnormalities encountered in human disease HPO. Added by default.
- Pathogenicity is a variant filter by pathogenicity determined manually by the user, by ACMG criteria or by pathogenicity database: Pathogenic, Likely pathogenic, Uncertain significance conflict (conflicting interpretations of pathogenicity), Uncertain significance, Likely benign, Benign, Undefined.
- Label is a variant filter by significance defined by the user: High significance, Moderate significance, Low significance, Sequencing error, Undefined.
- ClinVar is a variant filter by the clinical significance of the phenotype according to ClinVar database (you can find the description of values here).
- Consequence is a variant filter by effects on genes. The values are described here.
- Caller Filters is a variant filter by GATK variant filters.
- Min Depth level is a variant filtering scale by the minimum level of sequencing depth. Added by default.

- Trio GT is a variant filter by inheritance: De novo variants, Newly formed homozygotes (homozygotes in proband that are heterozygous in parents) or Single parent-inherited. Added by default, available only in Family group analysis for Trio case (Proband, Father and Mother).
- gnomAD 3 AF is a variant filter by allele frequency (AF) in gnomAD 3 database:
- <0.05 (threshold between common and rare variants) or empty (no allele frequency data in gnomAD);
- <0.02 (conditional threshold of a hypomorph) or empty;
- <0.01 (estimated threshold of autosomal recessive inheritance) or empty;
- <0.005 (estimated threshold of X-linked inheritance) or empty;
- <0.0001 (estimated threshold of autosomal dominant inheritance) or empty.
- Region - filter variants by the position on genome (chromosome, start and end positions).
Preferred gene transcript model#
On "Gene" tab of the variant details panel, gene transcripts from two databases are available: Ensembl and RefSeq. By default, a table with transcripts from Ensembl is shown.
In this section, you can configure which gene transcript model will be shown by default:

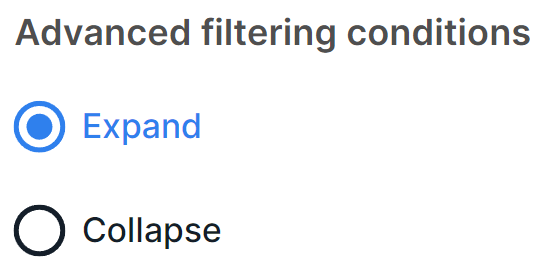
Advanced filtering conditions#
In advanced filtering mode, the query conditions for the searched variants are displayed at the top of the SNV Viewer table. When there are a lot of conditions in a query, it is convenient to collapse them so that they do not take up much space on the page.
In this section, you can configure whether the query conditions will be expanded (by default) or collapsed immediately after applying the filtering query:
SNV & CNV Viewer#
SNV Viewer is an embedded service for viewing and analyzing variants.
It is described in detail in the corresponding section.
CNV Viewer is an embedded service for viewing and analyzing copy number variations (CNVs).
Variant details bottom panel#
The variant detailed information panel contains main information about the selected variant. By default, it is expanded at the bottom of the SNV or CNV Viewer table. If you don't usually use the panel, you can hide it from SNV and CNV Viewers by selecting the "Collapse" option:

Return to default settings#
If you have made changes to the language selection, navigation bar, patient page, SNV Viewer, or
CNV Viewer settings, you can return to the default settings by
clicking on  . Please note that the action
will immediately reset all changes that you made on the page (except for changes to email, login and password).
. Please note that the action
will immediately reset all changes that you made on the page (except for changes to email, login and password).