Group Analysis
Group analysis is a combining of germline variants (SNVs/Indels) discovered in samples selected for analysis. Group analysis includes combining, filtration and annotation of germline variants of the samples.
Available types of group analysis:
- Germline cohort analysis is used to combine and compare samples that are related in some way (e.g. by pathology type or data source), or samples of large families in which sibling sequencing was performed. The analysis can include a maximum of 25 samples. The analysis results are presented in SNV Viewer with all variants discovered in the samples included in the analysis.
- Family analysis is used to combine and compare samples from the same family (proband + one or two parents or two parents without a proband). May include 2 or 3 samples. The analysis results are presented in SNV Viewer with all variants discovered in the samples included in the analysis. SNV Viewer has specialized filters based on genotypes of family members.
- Population analysis is aimed at studying the distribution of the allele frequencies in a certain population. It can include an unlimited number of samples. The analysis results are presented as VCF file with the allele frequencies of the variants discovered in the samples included in the analysis.
To open page with group analyses, open "All analyses" page from "Group Analysis" page block via the navigation panel on the left:
Create Group Analysis#
To start the group analysis creation,
click on .
Another way to create a group analysis is to consolidate a cohort, described here.
In the window that opens, enter a group analysis name in Name field. You can also add an analysis description here.
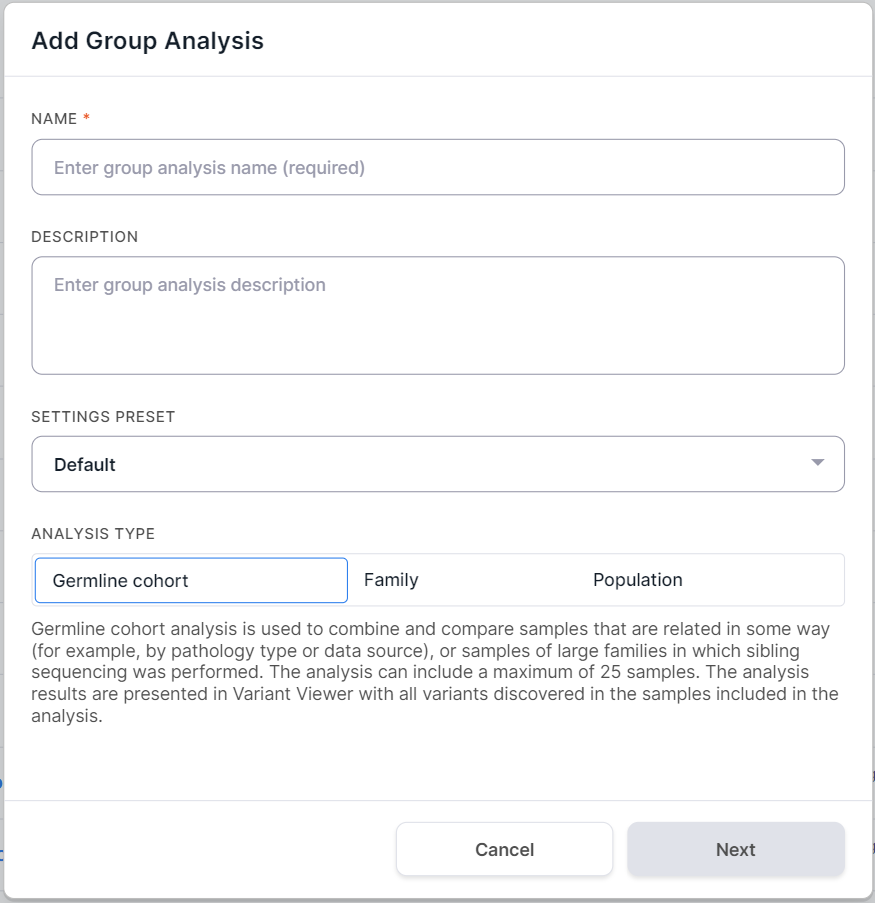
Settings Preset#
If you want to use your settings preset for analysis, select it from "Settings preset" drop-down list. Note that the group analysis can only be affected by a preset that configures germline SNVs/Indels filtration. If you are satisfied with the default variant filtration parameters, leave the default preset selected for analysis.
Analysis Type#
The available group analysis types are described above. This may be a germline cohort analysis, a family analysis,
or a population analysis. Select the analysis type by clicking on the corresponding container, and then
click to proceed to the selection of
samples for analysis.
Samples for Analysis#
The table with samples for which group analysis can be performed includes the sample name and the patient
code or run name to which it refers. A sample of
non-tumor tissue is marked as , a sample of tumor tissue - as
.
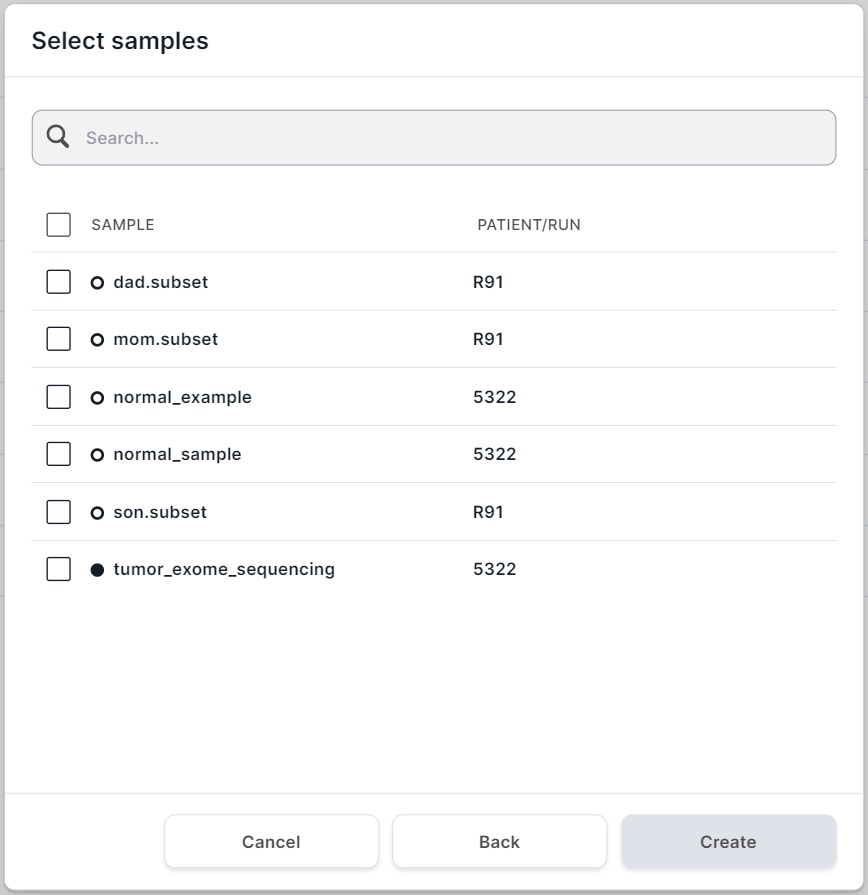
Conditions for samples that are available for group analysis:
- Samples were uploaded in FASTQ or BAM format;
- Germline SNVs/Indels discovery and annotation stages have successfully completed for samples;
- Samples refer to a patient or run that is not in "Archive".
Check the boxes of the samples (at least two) which you want to analyze. A maximum of 25 samples can be included in a germline cohort analysis, a maximum of 3 samples can be included in a family analysis, and an unlimited number of samples can be included in a population analysis. The names of the samples added to analysis must be unique. If you want to add the same samples to your analysis, rename one of them (you can do this on the sample page), so that the names are not repeated.
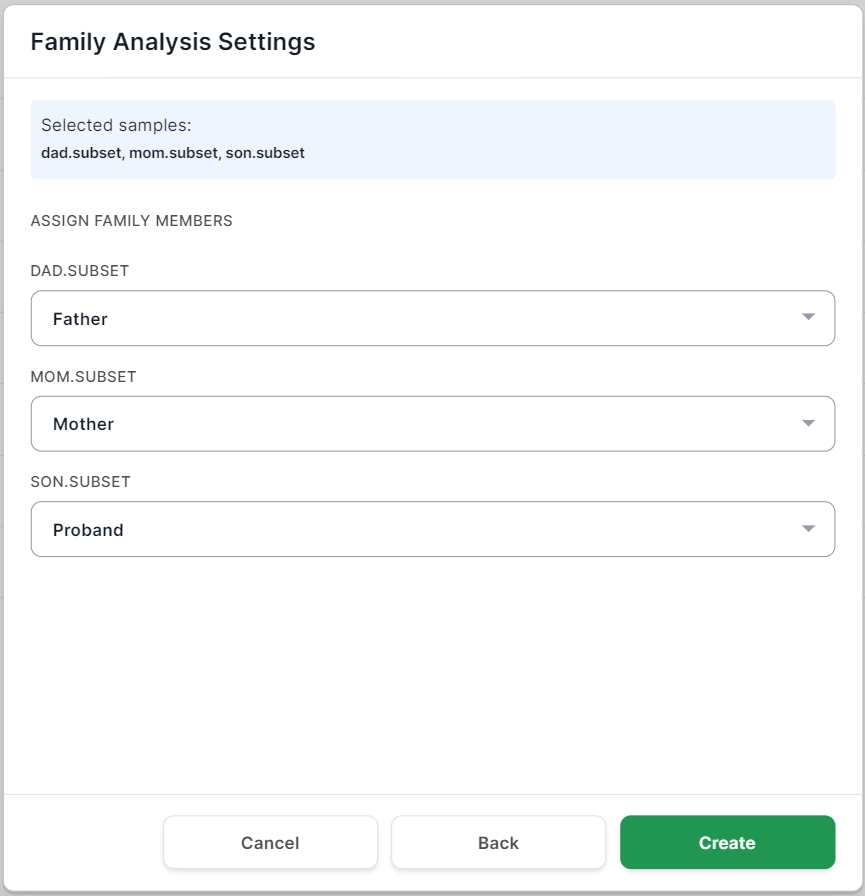
Family Analysis Settings#
If you have selected the family type of group analysis,
click on after selecting the
samples for analysis. You will see the family analysis settings page:
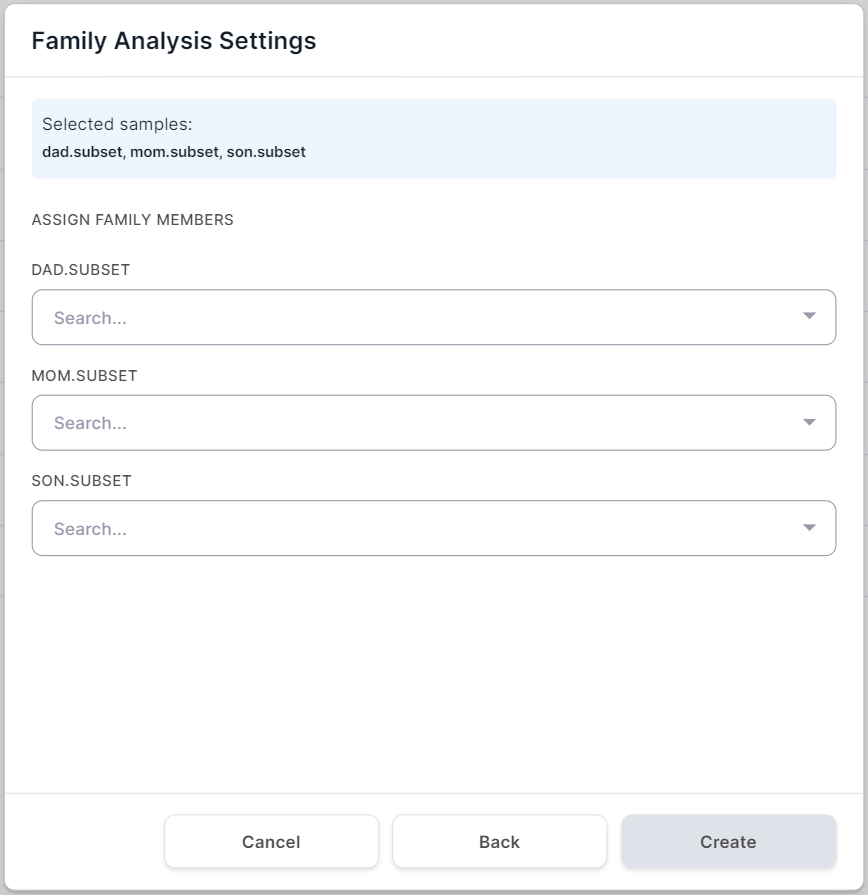
Define for each sample the "family role" of the patient from whom it was obtained: Proband (child), Father or Mother:
Run Analysis#
To start analysis, click on .
Group Analysis Page#
After starting the analysis, you will see its page ("Main" tab):
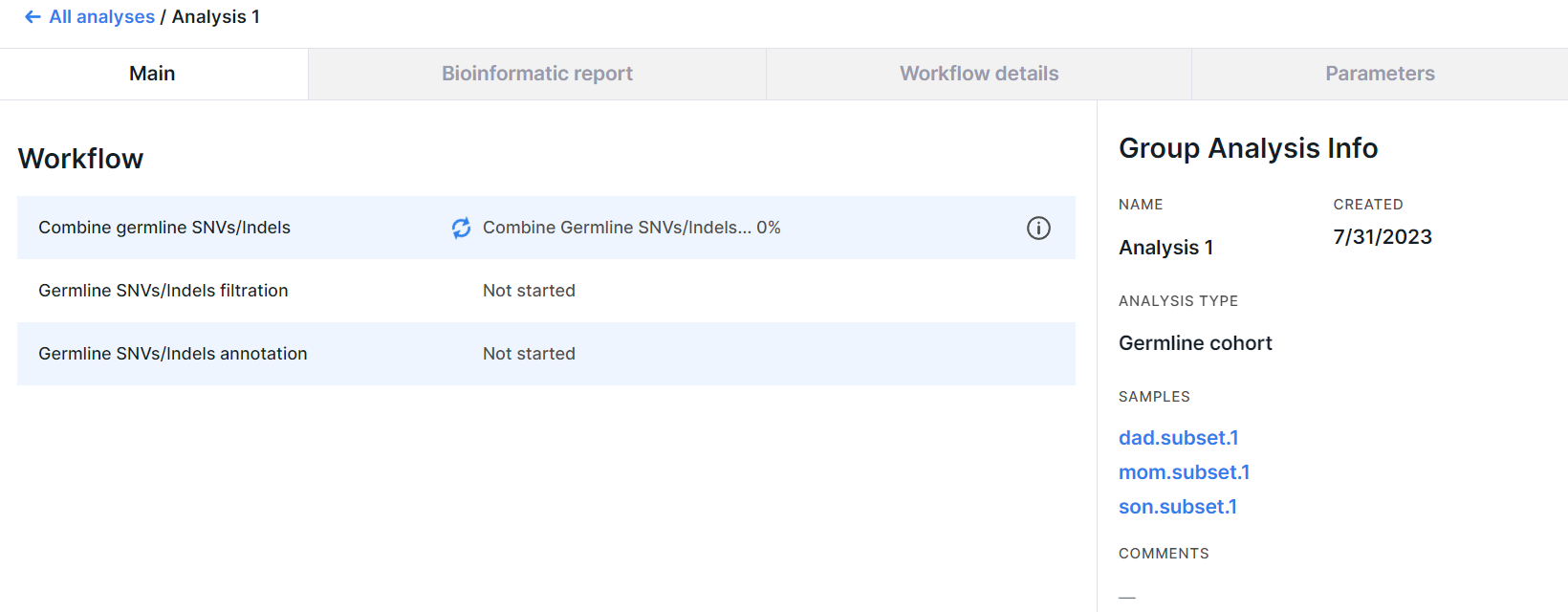
On the page, there is an information about the analysis and its workflow. When the analysis is successfully completed, the main results will appear here.
Group Analysis Info#
On the right side of the group analysis page, there is a basic information about the analysis:
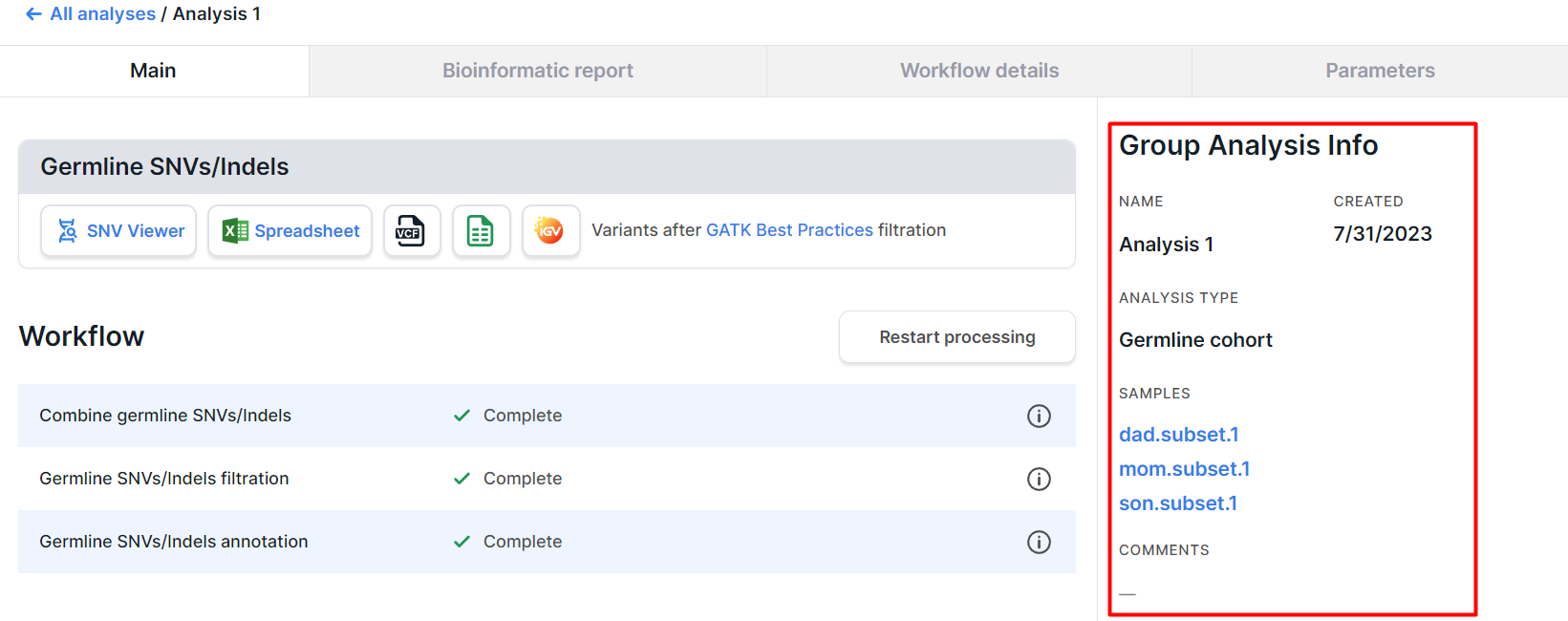
- NAME is the group analysis name. You can edit it by clicking on the value field.
- CREATED is the date when the group analysis was created in m/d/yyyy format.
- ANALYSIS TYPE: Germline cohort, Family, or Population. The types "Family" and "Germline cohort" can be changed between each other by clicking on the value field and selecting a different type of analysis (if the samples included in analysis have not been deleted).
- SAMPLES is a list of samples for which the analysis was carried out. By clicking on the sample name, you can open its page. If a family analysis was performed, each sample will also be assigned to the "family role" of the patient from whom it was obtained:

- COMMENTS: can be entered by clicking on the value field.
Analysis Workflow#
On the left side of the group analysis page, there is an information about the analysis workflow. The information is presented in the form of a table with analysis stages and their statuses:
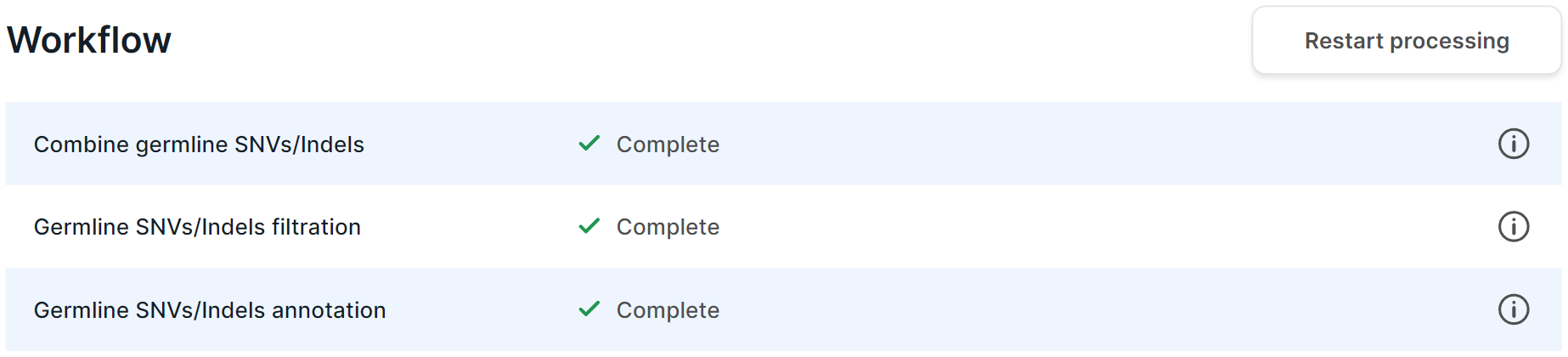
- If the stage is in progress, then
its status has
icon and a percentage value of its progress.
- If the stage is completed successfully,
then it has the status
.
- If the stage is completed with an error,
then its status has
icon and the failed task is specified. In this case, you can report a problem to the administrator by clicking on
, or retry the task by clicking on
.
- If the stage is included in the workflow, but the necessary previous steps of the analysis have not yet been completed, then it has the "Not started" status.
You can view the details of each stage by
clicking on .
Then you will see this stage section on "Workflow details" tab.
On this tab, for each stage, you can download the result files and view technical details, such as console
output and processing error message (if an error occurred during the analysis).
If you would like to receive notifications when the group analysis processing is complete, please enable the corresponding option in your account settings.
Group Analysis Stages:#
- Combine Germline SNVs/Indels: merging gVCF files resulting from germline SNVs/Indels discovery in samples using GATK CombineGVCFs tool, and joint genotyping of analysis samples using GATK GenotypeGVCFs;
- Filter Germline SNVs/Indels: filtering variants using GATK VariantFiltration tool based on certain filtering criteria that you can find and change on "Parameters" tab of the group analysis page;
- Annotate Germline SNVs/Indels: determination of the variant impact on DNA regions using VEP tool and a set of tools developed by Genomenal;
- Store annotated variants for SNV Viewer: no such stage for population analysis.
Restart Processing#
If you want to restart the analysis from one of the completed stages, hover over the stage row and
click on . If you want to restart the analysis from scratch (from "Combine Germline SNVs/Indels" stage), click on
.
Please note that this button is available only for analysis, all stages of which were completed successfully.
Analysis Results#
After all stages of analysis are completed successfully, the results will appear on the group analysis page:
- "Germline SNVs/Indels" section on "Main" tab including all the main results of group analysis. For germline cohort and family analyses, the results look like this (includes SNV Viewer):

And for the population analysis, the results look like this (no SNV Viewer included):

- SNV Viewer is a embedded service for viewing and analyzing variants.
You can read more about it below.
SNV Viewer is not provided for population analysis.
Click on
 to open SNV Viewer.
to open SNV Viewer. - Text file in CSV (Comma-Separated Values) format with annotated variants. To download the file,
click on
.
- File in VCF 4.2 (Variant Call Format) format with
annotated variants.
For germline cohort and family analyses, click on
to download file. For population analysis, click on
to open a drop-down list with result files:
- Frequency is a VCF file with the allele frequencies of the variants discovered in the samples included in the analysis.
- Annotated is a VCF file with annotated variants.
- Annotated variants in Google Spreadsheet (only a limited number of variants can be shown). To open a table,
click on
.
- Integrative Genomics Viewer (IGV) is an embedded
module for visualization of variants on the genome. To open the module,
click on
.
Germline SNVs/Indels Report for Group of Samples on "Bioinformatic report" tab. The report includes information on the count, genome position, and representation in databases of germline variants resulting of the analysis. A detailed description of the report can be found here. To export the report in PDF format, click on
.
Result files for each stage of the analysis on "Workflow details" tab.
Group Analysis SNV Viewer#
SNV Viewer is only provided for germline cohort and family analyses. It is described in detail in the corresponding section on Sample SNV Viewer, however, Group Analysis SNV Viewer has a number of special features:
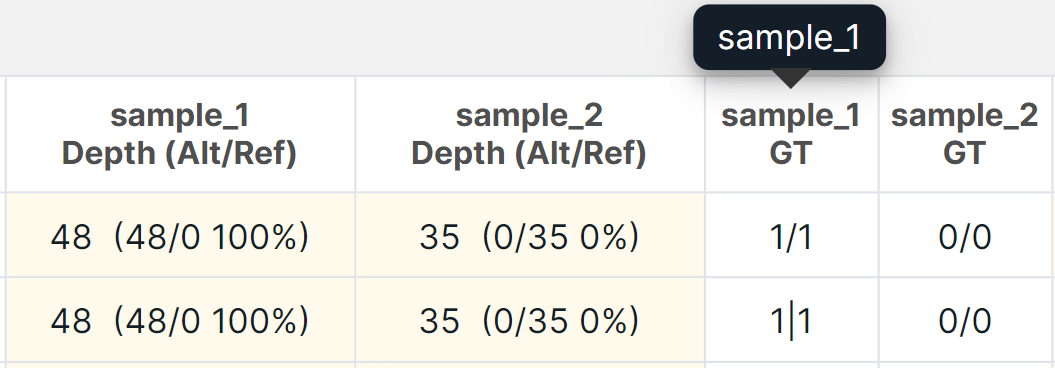
- The columns "Depth (Alt/Ref)" and "GT" contain the data of all samples of the analysis. For germline cohort analysis, these column names contains the name of the sample, in which the variant has the corresponding sequencing depth and genotype (when you hover over the column name, you can see the full sample name):
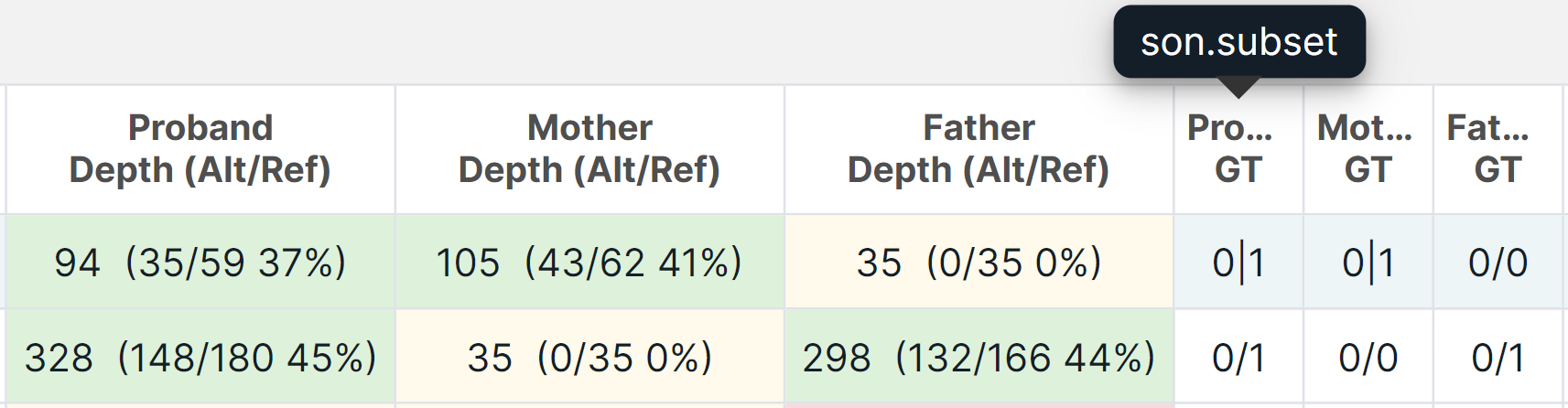
For family analysis, samples in which a variant has the corresponding sequencing depth and genotype are designated by certain patient "family roles" (when you hover over the column name, you can see the sample name):
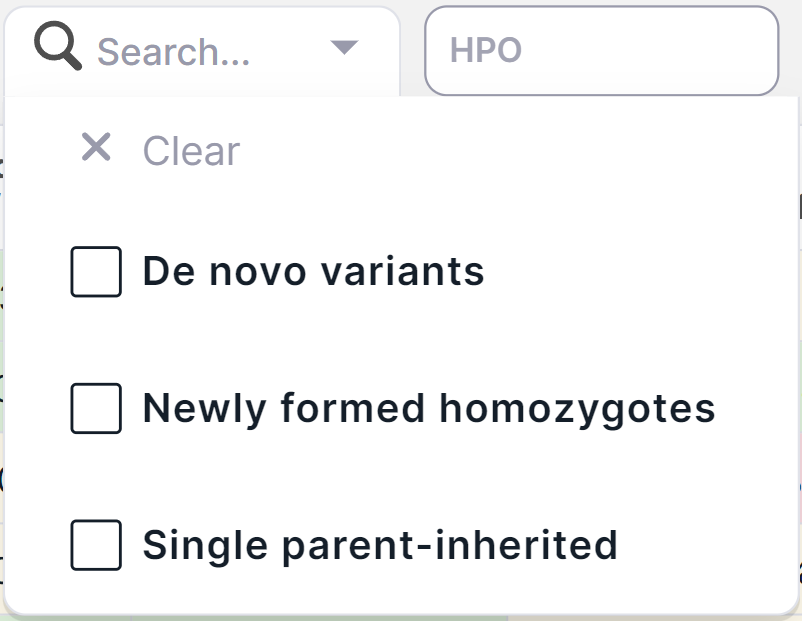
- The basic filter "Trio GT" is a filter of variants by inheritance: De novo variants, Newly formed
homozygotes (homozygotes for loci in the proband that are heterozygous in parents) or Single-parent inherited.
Available by default only in Trio case of family analysis (Proband, Father
and Mother). To filter variants by inheritance, click on the filter
window
and tick the required values in the drop-down list:
To clear the filter by inheritance, click on the filter window,
and then on .
You can also filter by trio GT in advanced filtering mode using "Trio GT (Genotype)" filter.
- There is no column, basic filter and advanced filter "Origin", since all variants in group analysis SNV Viewer are germline.
- There is no button to include variants to report, because no reports are generated for group analysis.
- The variant pathogenicity can be determined only manually or by pathogenicity base.
- On the variant details page, there are only "Annotation" and "Occurrences" tabs.
Archive, Restore and Delete Group Analysis#
To archive group analysis, hover over the group analysis row on "All analyses" page and
click on . The analysis will be moved to
"Archive" page from "Group Analysis" page block. On "Archive" page, you can:
- Restore group analysis by hovering over the group analysis row and
clicking on
. The analysis will then be moved back to "All analyses" page.
- Delete group analysis by hovering over the group analysis row and
clicking on
. Then you will see a confirmation window for deleting the analysis:
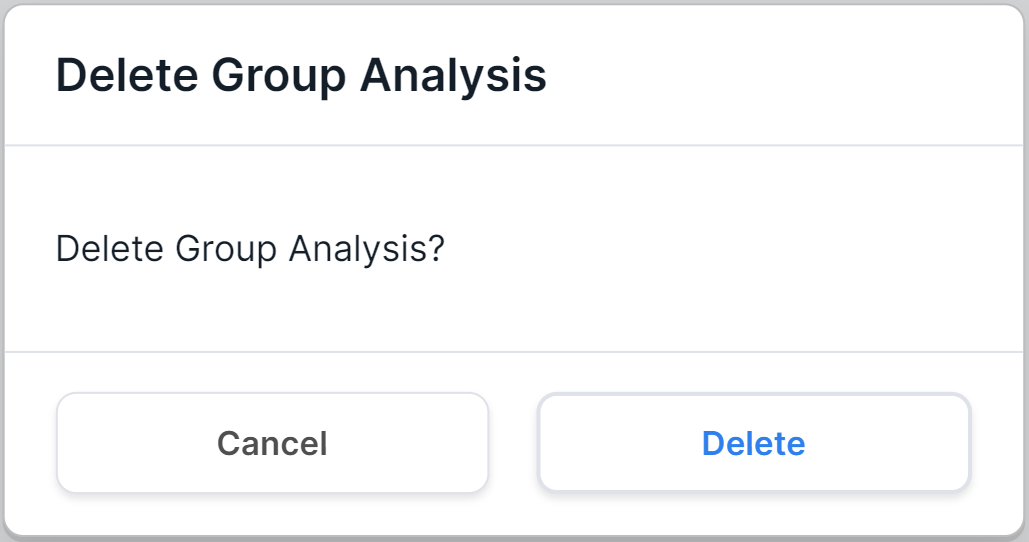
To confirm the analysis deletion, click on . To cancel deleting an analysis, click on
or outside the confirmation window.
In addition, you can delete all archived group analyses by
clicking on  .
.