Samples Upload Page
On the samples upload page, you can perform multiple uploads of data in different patients.
It can be useful if you have a research purpose and do not care which patient the sequencing samples
came from. To open the samples upload page, click on
the button on the all patients page.
You will see the following page:
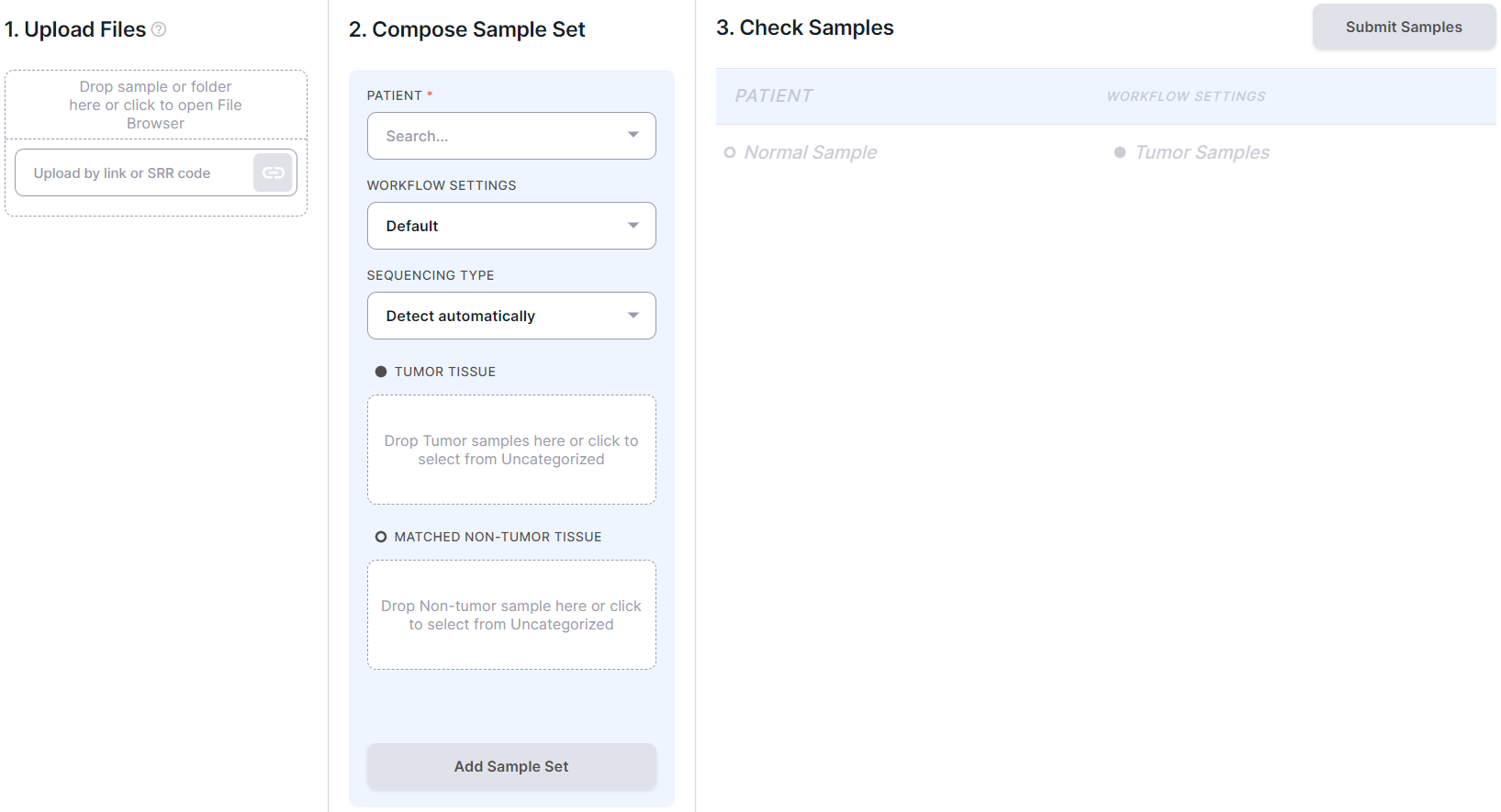
- Upload samples files. Supported data formats are described here.
The sample files can be uploaded:
- From computer
- From FTP, HTTP or Yandex.Disk servers
- From Google Drive
- From NCBI SRA database
To upload, drag and drop files or folder containing files into the drop-area or click on it to open the File Browser:
To upload, paste the link to the file or folder containing files on the server into the placeholder:
and click on the button or press Enter on the keyboard.
To upload a file from Google Drive, first share the file with anyone with the link (how to do this is described here).
Paste the resulting link into the window:
and click on the button or press Enter on the keyboard.
To upload, paste the run code from NCBI SRA (e.g. SRR24755541) into the placeholder:
and click on the button or press Enter on the keyboard.
Samples uploaded by mistake can be deleted by hovering over their card and
clicking on .
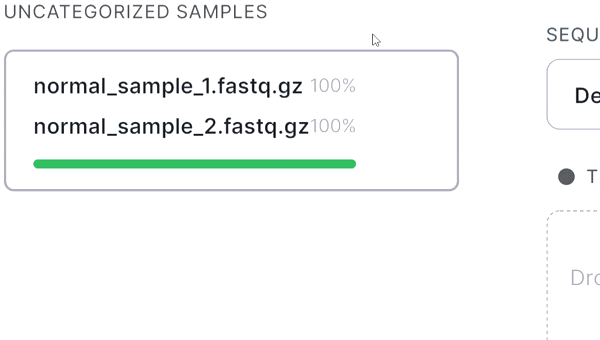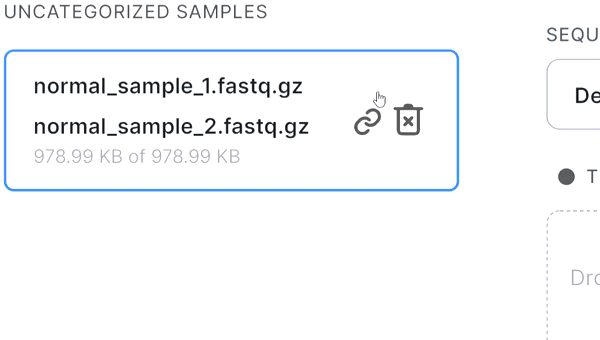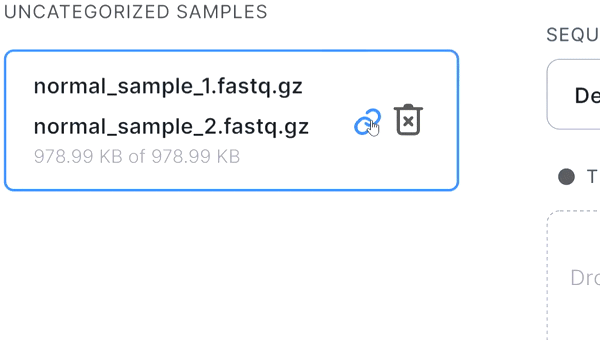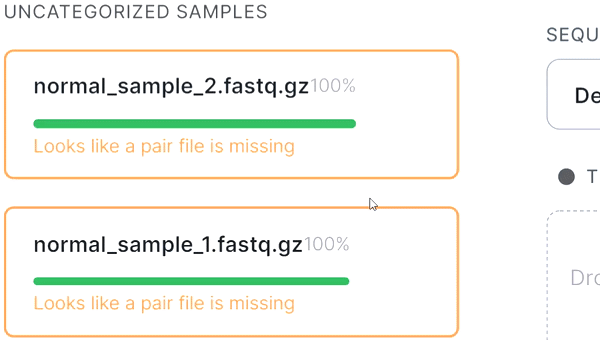
Recognizing paired files#
- If your sequencing is paired-end and a pair of FASTQ files of the same sample is named in an Illumina convention (or even by some other common read naming scheme), the files will be automatically recognized as a pair when added:
- If the pair is not recognized correctly, you can manually unlink files by hovering over the pair and
clicking on
:
- If the automatic pair recognition did nоt succeed, you can manually pair the files. To do this, either drag the mate paired-end file to the primary one:
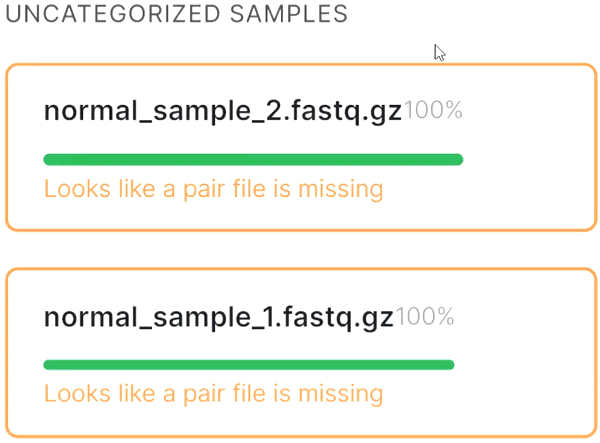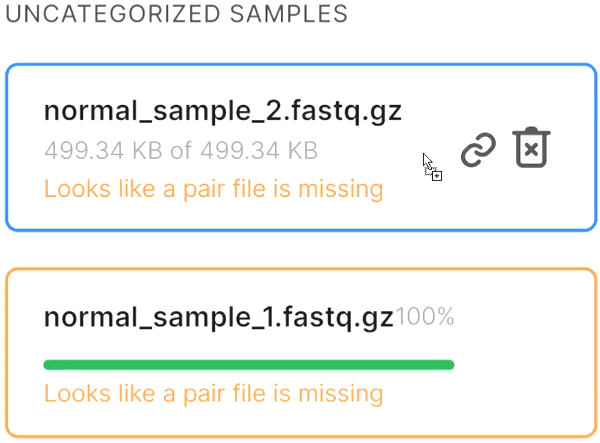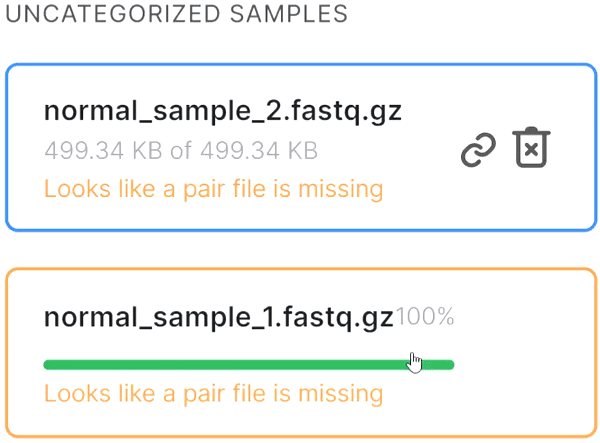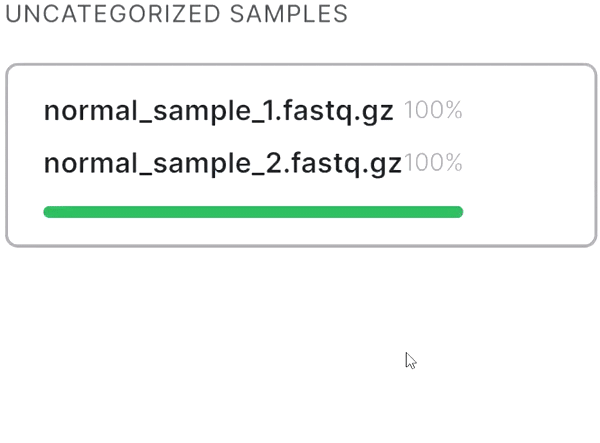
or click on the paperclip in the primary file card and then click on the mate file:

Merging sequencing lanes#
- If your sequencing sample consists of multiple FASTQ files that correspond to different lanes on the
flow cell and are named the same except for the sequencing lane designation:
_L[lane number](e.g. "_L01"), then the lane files will automatically be merged into one sample. In this case, pairing will be taken into account for the lanes in the same way as described above, i.e. paired lane files will be merged into a paired sample, and unpaired lane files will be merged into an unpaired sample.
An example of merging four paired lanes into one sample:
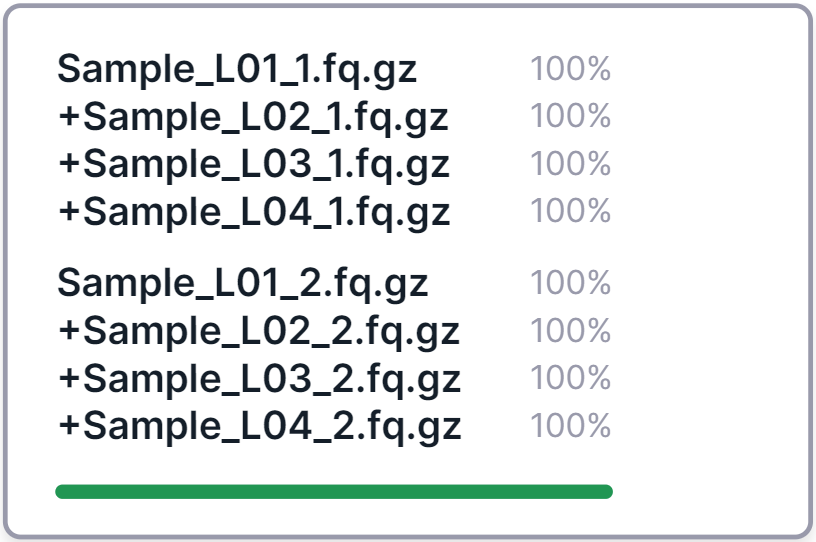
The first file in the merged sample card is the one with the lowest lane number among the others, and the remaining lanes are added to it, symbolized by a plus in front of their names.
- If the files are merged incorrectly, you can split them by clicking on
the button
 , which is located under all
uploaded files in the "Uncategorized samples" section.
, which is located under all
uploaded files in the "Uncategorized samples" section. - If the necessary files are not merged into one sample, then you can merge them manually, either by
dragging the card of one lane file to another (to merge these two files), or by clicking on
the button
 , which is located under all
uploaded files (to merge all uploaded files). Note that the button will be available only if there are samples
of the same pairing in the "Uncategorized samples" section (i.e. either all samples are paired or all samples
are unpaired).
, which is located under all
uploaded files (to merge all uploaded files). Note that the button will be available only if there are samples
of the same pairing in the "Uncategorized samples" section (i.e. either all samples are paired or all samples
are unpaired).
note
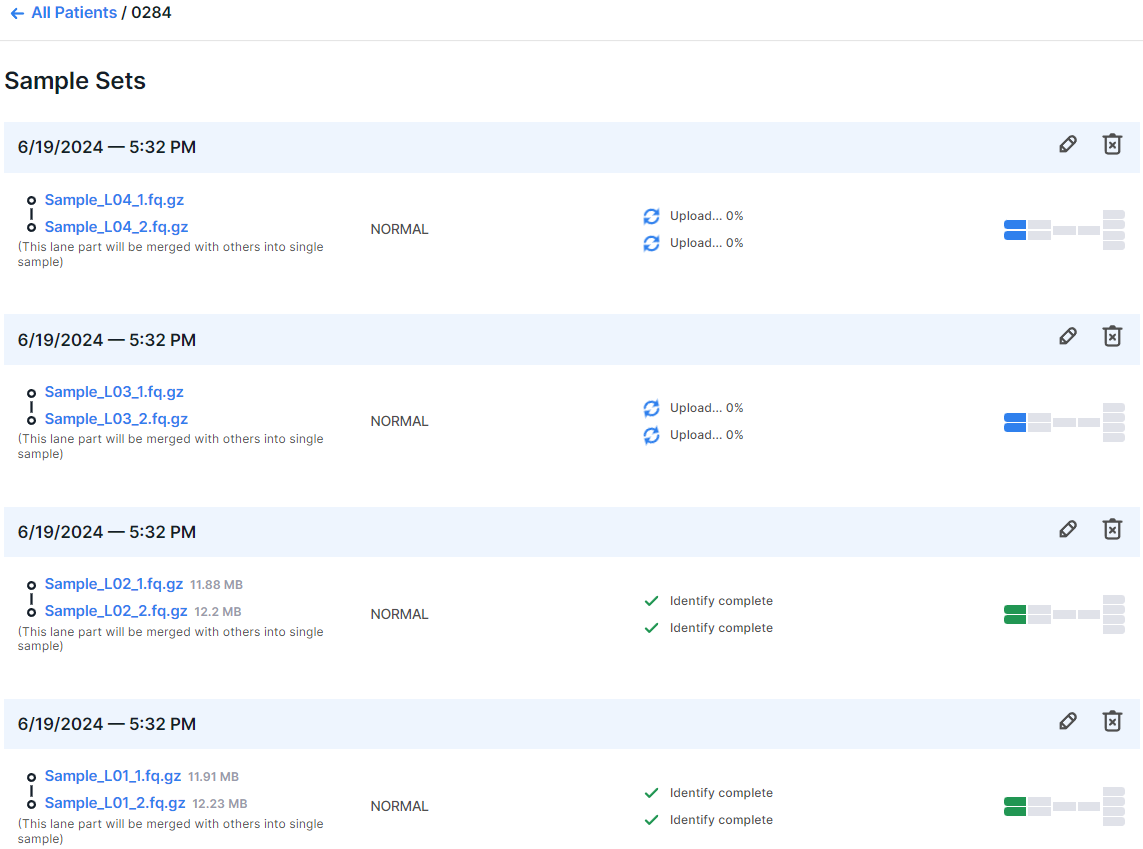
After running the analysis, a sample consisting of multiple lanes will initially appear as multiple samples. At the bottom of each "sample" (which is actually a part of the future sample) it will be indicated that "This lane part will be merged with others into single sample". The merging of lanes into one sample will occur after the successful completion of the uploading and identification stages for all the lanes of the sample.
- Select workflow settings:
- for a pair of tumor/normal samples: if you want to analyze a tumor sample to look for somatic and germline mutations in contrast with a normal sample, select Somatic and germline variant analysis workflow setting;
- for tumor tissue samples: if you want to analyze a tumor sample to look for somatic and germline mutations without a normal sample, select Somatic and germline variant analysis workflow setting;
- for normal samples: if you want to analyze a normal sample to look for germline mutations, structural and copy number variations, select Germline variant analysis workflow setting, and if for prenatal genetic testing (copy number variations discovery), select Prenatal analysis workflow setting.
- Compose sample set: drag the uploaded samples (the "Uncategorized samples" section on the left)
into the sample placeholders of the corresponding type, or click on the uploaded sample card and then on
the corresponding placeholder.
If you select "Somatic and germline variant analysis" as the workflow setting, then two sample placeholders are available: "Tumor tissue" and "Matched non-tumor tissue", which are suitable for tumor tissue samples and non-tumor tissue samples, respectively. You can define an unlimited number of samples as tumor (after defining the first sample, another placeholder for tumor tissue samples appears) and only one sample as non-tumor.
If you select "Germline variant analysis" or "Prenatal analysis" as the workflow setting, then only one placeholder will be available, which is only suitable for normal samples:

You can define an unlimited number of samples as normal: after defining the first sample, another placeholder appears.
If you misplaced sample, you can drag it to the sample placeholder of a different type, or return it to the
"Uncategorized samples" section by hovering over the sample and clicking on
the cross .
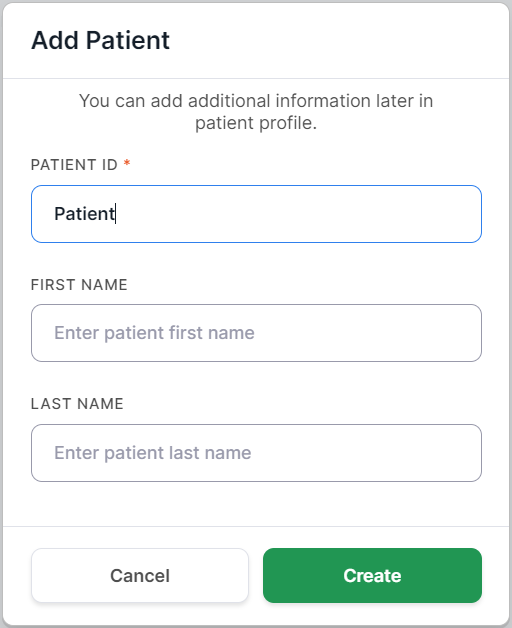
- Link a sample set to a patient: select an already added patient from the list or
link a set to a new patient - to do this, open the drop-down list in the "Patient" field and select the
option
. You will see the form for adding a new patient to the system. Only the Patient ID field is required. You can fill out the details later on this patient's personal page. Click on
to proceed:
- Define the sequencing type. The sequencing type is usually detected automatically by Genomenal.
However, if you are sure which sequencing type was used to obtain your data, select the appropriate type
to speed up the analysis. This can be whole genome sequencing (WGS)
or targeted selection (sequencing with a targeted panel or whole-exome sequencing (WES)).
After selecting "Targeted selection" as the sequencing type, you will see an additional field "Capture kit features". The capture kit used during sequencing can be detected automatically by Genomenal. But if you are sure which targeted panel was used when obtaining your data, you can select it from the list of built-in or already uploaded panels in the system, as well as upload the appropriate capture kit by opening the drop-down list in the "Capture kit features" field and selecting the option. Then you will see a field with the choice of a reference genome for the capture kit. By default, the reference genome assembly corresponding to the genomic intervals of the uploaded capture kit is detected automatically by Genomenal. You can determine the assembly by clicking on the appropriate field and selecting the assembly in the drop-down list: hg19 or hg38. Then click on the button
 and
select the panel file in the opened File Browser.
and
select the panel file in the opened File Browser.
Please note that by selecting the sequencing type and capture kit, you define them for all samples of set at once. - Add sample set by clicking on
.
- Check sample sets: composed and added sample sets will be displayed on the right:

For each sample set, the patient, the workflow setting, and the samples included in the set are listed.
The non-tumor sample files are located in the left column of the set and are
marked as . The tumor sample files are located in the right column of the set and are marked as
. A pair of non-tumor sample files:
, a pair of tumor sample files:
.
You can edit the sample set by clicking on
the button . The samples will then return to the "2. Compose Sample Set" section. In addition, you can delete the sample set by clicking on the button
. Then the samples
will return to the "Uncategorized samples" section.
- Run samples analysis by clicking on
. The samples upload page will close and you will see the all patients page. To see the added sample set, open the page of the corresponding patient with whom you have linked the set.
Attention!
If you're uploading sample files from a computer, and not by a link, do not refresh or close the tab where files are being uploaded until it's complete. Otherwise, the upload will be interrupted. To resume an interrupted upload, you can use the form described in the corresponding section. After the upload is completed, you can close the tab, browser and turn off the computer - further analysis does not depend on your device.
You can track the sample processing on its page or through notifications about its processing completion.