Main
The "Main" tab on the sample page contains the following information:
1. Interpretation Results#
The section may include potential findings if at least one report, that uses potential findings, was generated for the sample, and interpretation results if sample interpretation was completed.

Potential Findings#
Potential findings are SNVs/Indels or copy number variations (CNVs) discovered in a sample that meet some predefined criteria. These may be ACMG criteria, filters for gene panels with additional conditions for rare monogenic diseases, etc.
Potential findings can include SNVs/Indels or CNVs from report blocks of the type “SNVs/Indels in ACMG SF genes” (“ACMG Secondary Findings Report”), “SNVs/Indels suitable for the specified conditions” or “CNVs suitable for the specified conditions". To have the listed blocks use the found mutations as potential findings, turn on the corresponding toggle in these blocks. The potential findings section will not appear on the sample page if no report, that uses potential findings, has been generated for the sample.
The potential findings section provides information on each generated report that uses potential findings. For each report, the number of findings discovered in it (SNVs/Indels or CNVs) is indicated. By clicking on a line with information for a specific report, you can open its page. In the report, you can change the status of a potential finding, i.e. include it to the interpretation results or exclude it from the list of potential findings. In both cases, it will no longer count towards the total number of potential findings in this report on the sample page. Once the status of all potential findings in a report has been determined, the message for that report will change to "No potential findings were found".
Interpretation Results#
Interpretation results are information about the number of mutations with a certain pathogenicity class discovered in the sample and included to the interpretation results. The section may include information about SNVs/Indels, designated "SNV", and information about copy number variations, designated "CNV". The information is presented in the form of pathogenicity icons indicating the number of mutations with this pathogenicity:
- pathogenic mutations:
;
- likely pathogenic mutations:
;
- mutations of uncertain significance:
;
- likely benign mutations:
;
- benign mutations:
.
If sample interpretation has not been completed, there will be no interpretation results section on the sample page. If sample interpretation has been completed, but mutations with a certain pathogenicity class have not been included to the interpretation results, then the result will be “No pathogenic variants found”.
2. The analysis results#
The section contains the main results of the analysis: a separate section with the results corresponds to a separate stage of the analysis. The availability of the results of a particular stage depends on whether it was carried out and completed successfully.
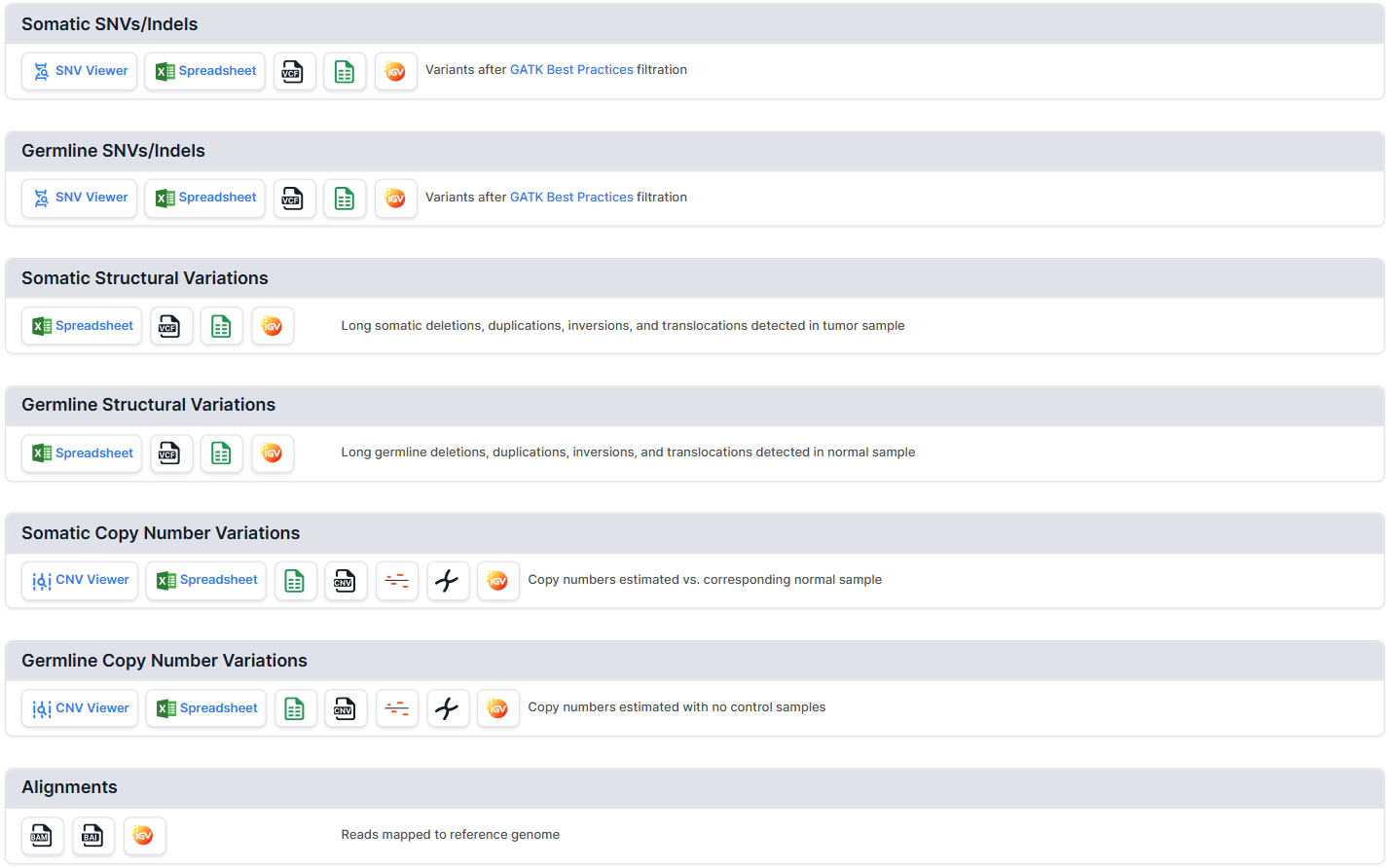
You can read more about each section in the corresponding section of the documentation:
- Somatic and Germline SNVs/Indels
- Somatic and Germline Structural Variations
- Somatic and Germline Copy Number Variations
- Alignments
3. Sample Info#
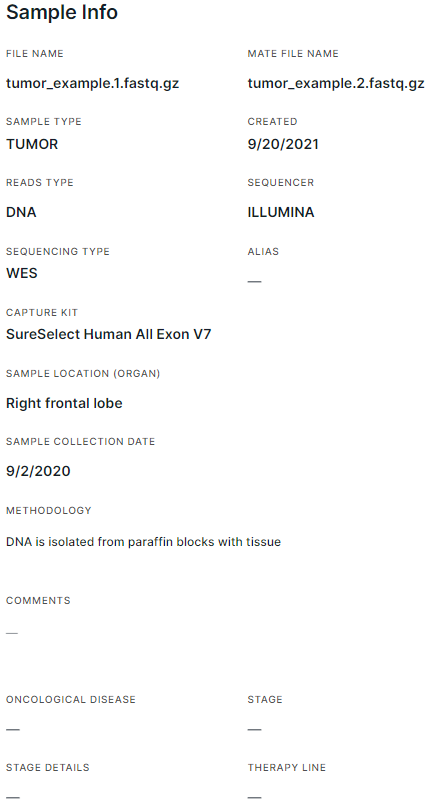
- FILE NAME and MATE FILE NAME are the names of the uploaded sample files with extensions (can be edited by clicking on the name).
- SAMPLE TYPE is the type that was selected when uploading the sample (TUMOR or NORMAL). You can change the type only by dragging the sample to the appropriate type in the edit sample set window, or by selecting a different type in the sample row (for runs).
- CREATED is the date of creation of a sample set (start of uploading sample files into the system) in the format M/d/yyyy.
- READS TYPE is the sequence type (DNA, RNA or UNKNOWN) that was sequenced. Determined by a built-in classifier that predicts the reads type using the LSTM (long short-term memory) model. If the type is not defined correctly, you can change it by clicking on it and selecting another type from the drop-down list.
- SEQUENCER is the type of sequencer on which the sequence was sequenced (ILLUMINA, ION TORRENT, BGI or UNKNOWN). The type is determined according to the ID of the reads from the file in FASTQ format. If a sequencer is not correctly defined, it can be changed by clicking on it and selecting a different type from the drop-down list.
- SEQUENCING TYPE: Panel (targeted sequencing), WES (whole-exome sequencing), WGS (whole-genome sequencing), Low-pass WGS (whole-genome sequencing with low coverage), UNKNOWN (the sequencing type may not be detected for some samples in VCF format). Detected automatically by Genomenal, may be based on Sequencing Type and Capture Kit selected while composing sample set. The type can be changed on "Parameters" tab ("Sequencing Type" parameter: "WGS" option corresponds to either type "WGS" or "Low-pass WGS", "Targeted selection" option corresponds to "Panel" or "WES" type).
- ALIAS is a synonymous name for the sample, which can be entered by clicking on the value field.
- CAPTURE KIT is a panel used in targeted selection. Detected automatically by Genomenal, or by user while adding sample set. If the capture kit is defined incorrectly, it can be changed on Parameters tab. You can use both built-in capture kits (standard panels often used in sequencing) and custom capture kits uploaded by the user (on "Capture kits" page).
- SAMPLE LOCATION (ORGAN) is the organism part from which the sample was taken for sequencing. Can be specified by clicking on the value field.
- SAMPLE COLLECTION DATE is the date in the format M/d/yyyy. Can be specified by clicking on the value field.
- METHODOLOGY is the method of extracting the sample. Can be specified by clicking on the value field.
- COMMENTS - can be specified by clicking on the value field.
- ONCOLOGICAL DISEASE, STAGE and STAGE DETAILS (fields are presented only on the tumor sample page) are the patient's oncological disease, its stage and stage details, which were defined for the patient at the time the sample was taken for sequencing.
- THERAPY LINE (field is presented only on the tumor sample page) is the therapy line that was being used to treat the patient's disease at the time the sample was taken for sequencing.
4. Workflow#
The section is a table with analysis stages and their statuses.
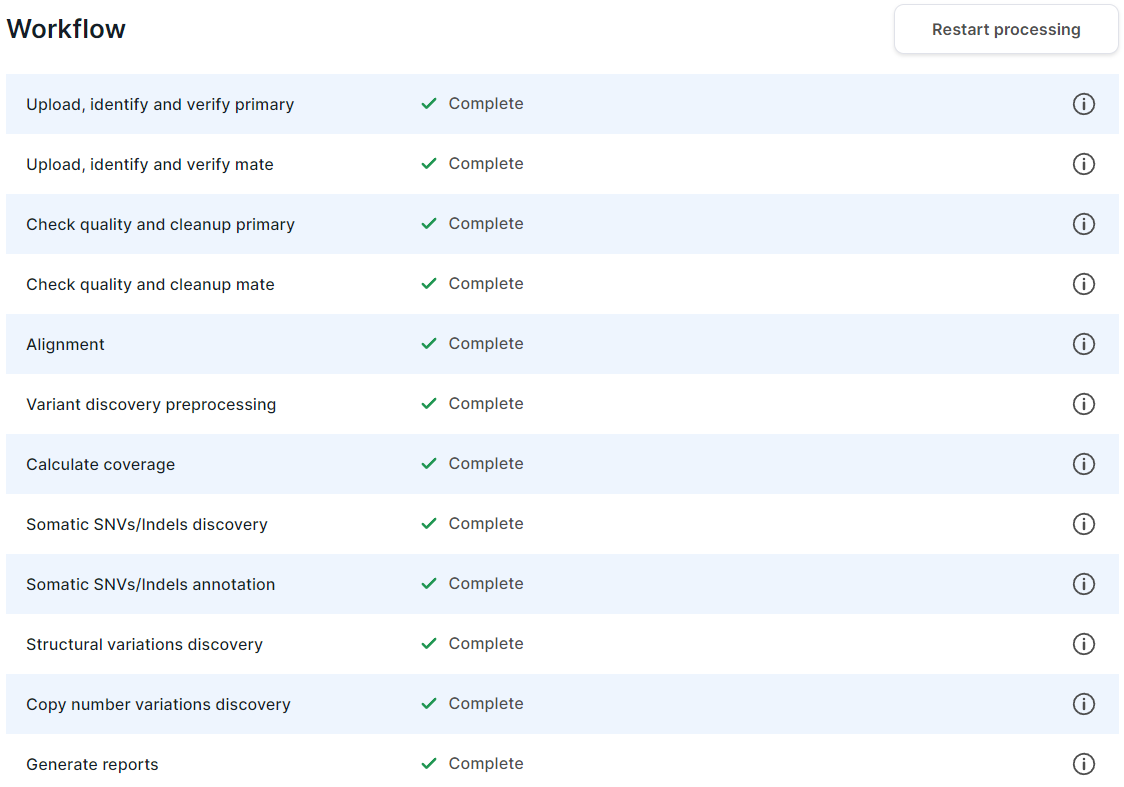
- If the stage is in progress, then
its status has
icon and a percentage value of its progress.
- If the stage is queued or waiting for the prerequisite stages of the control sample to be
completed (in the case of the tumor sample from a pair of tumor/normal samples), then
its status has
icon.
- If the stage is completed successfully,
then it has the status
.
- If the stage is completed successfully with non-critical problems,
then it has the status
and indicates these problems.
- If the stage is completed with an error,
then its status has
icon and the failed task is specified. In this case, you can report a problem to the administrator by clicking on
, or retry the task by clicking on
.
- If the stage is included in the workflow, but the necessary previous steps of the analysis have not yet been completed, then it has the "Not started" status.
- If the stage is excluded from the analysis (in the workflow settings used when adding sample set or on the "Parameters" tab of the sample), then it has the "Turned off" status.
You can view the details of each stage by
clicking on .
Then you will see this stage section on "Workflow details" tab.
If you want to restart the analysis from the very beginning (not counting the uploading of the sample files),
click on . Please note that this button is available only for those samples, all stages of analysis of which were completed successfully. You can also restart the analysis from a certain stage by hovering over its row and clicking on
.