Glioma Report
Glioma report is a clinical report on the characteristics of gliomas (including recurrent glioblastoma) based on the onco-markers discovered in samples and a base built on the basis of expert knowledge and scientific publications on this topic.
caution
The results of the report should be interpreted using all available clinical and laboratory data and should not be used in isolation for diagnosis and treatment.
Glioma report is generated for the sample if the following conditions are met:
- The sample is uploaded as a tumor tissue sample.
- The sample analysis was successfully completed (that is, all stages of the analysis have the status "Complete").
- For a single tumor sample (tumor-only), at least one of the following stages was successfully completed: "Somatic SNVs/Indels annotation", "Germline SNVs/Indels annotation", or "Copy number variations discovery". If you have a set of tumor and corresponding non-tumor samples (tumor-normal), then it is necessary that the "Somatic SNVs/Indels annotation" stage was successfully completed for tumor sample, and "Germline SNVs/Indels annotation" stage was successfully completed for normal sample.
- The corresponding report template is created (before the sample was processed) and active.
Applicability to Analysis of Other Pathologies#
The report contains both information specific to gliomas and sections that can be used to present the analysis results of other pathologies.
Sections specific to gliomas:
- Tier I: Variants of Strong Clinical Significance;
- Tier II: Variants of Potential Clinical Significance;
- Suggested diagnosis;
- Suggested drugs;
- Mutations details;
- Tier III: Mutations in other Cancer-panel genes;
- Glioma genes panel details;
- Pathogenic or likely pathogenic for gliomas;
- Pathogenic or likely pathogenic for any other genes/diseases;
- Glioma associated;
- Reference list.
Sections not specific to gliomas:
- Patient info;
- Tumor sample info;
- Immunotherapy markers;
- Clinical trials;
- Control sample info;
- ACMG secondary findings;
- Technical limitations.
Patient Info#
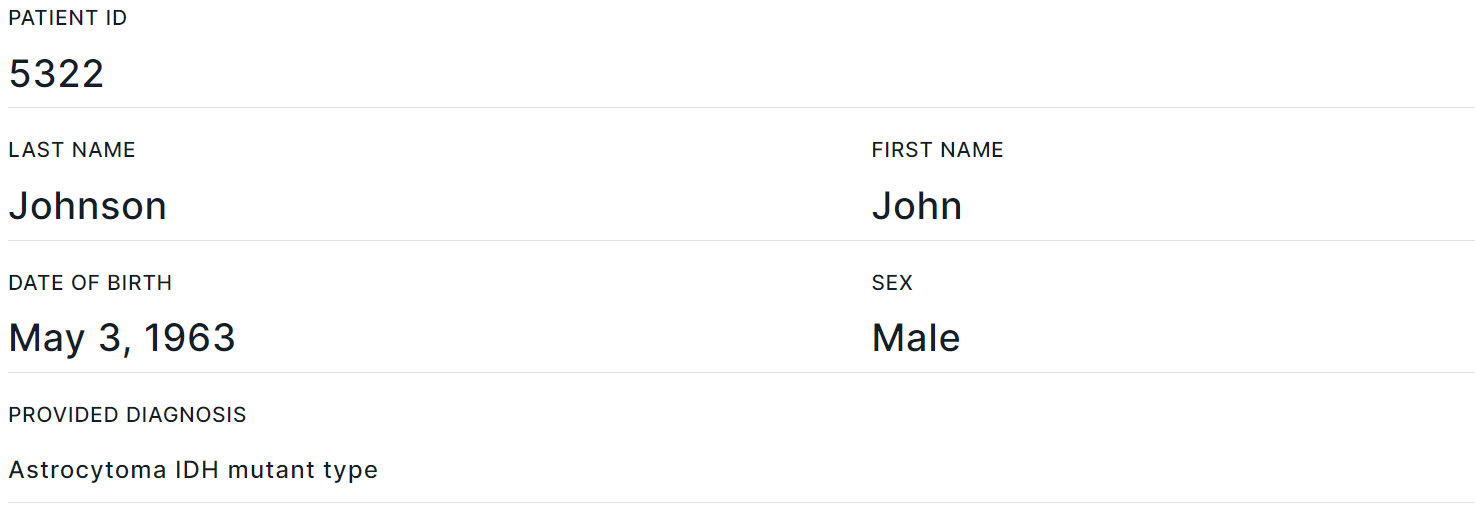
- Corresponds to "Patient info" template block.
- Contains basic data about the patient: Patient ID, Last name, First name, Date of birth, Sex, Diagnosis. You can also add fields with patient Middle name, Age, Date of diagnosis and Comments in the report template.
- Section fields can be filled in or edited on the report page, on the patient page, or on "Sample" tab of the variant details page. Information is updated in the report automatically, regardless of where the data was updated.
1. Somatic Mutations Report#
- This section contains information obtained from the tumor sample analysis.
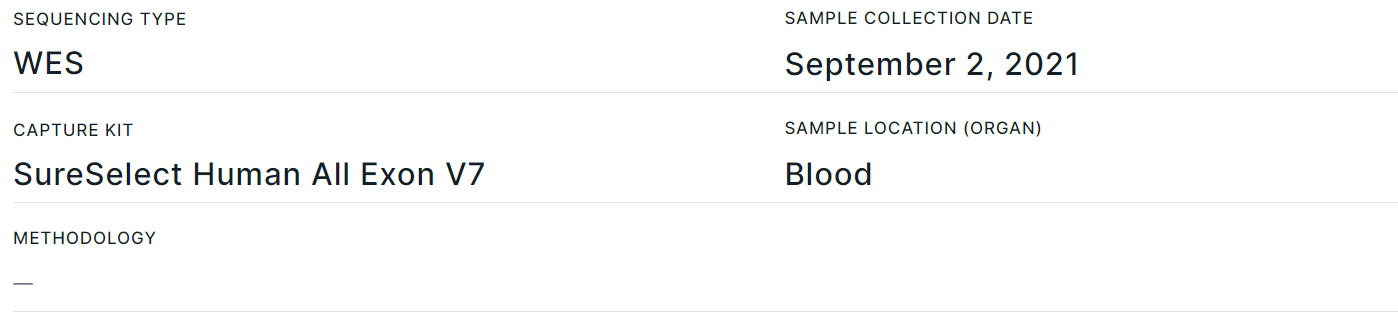
Tumor Sample Info#
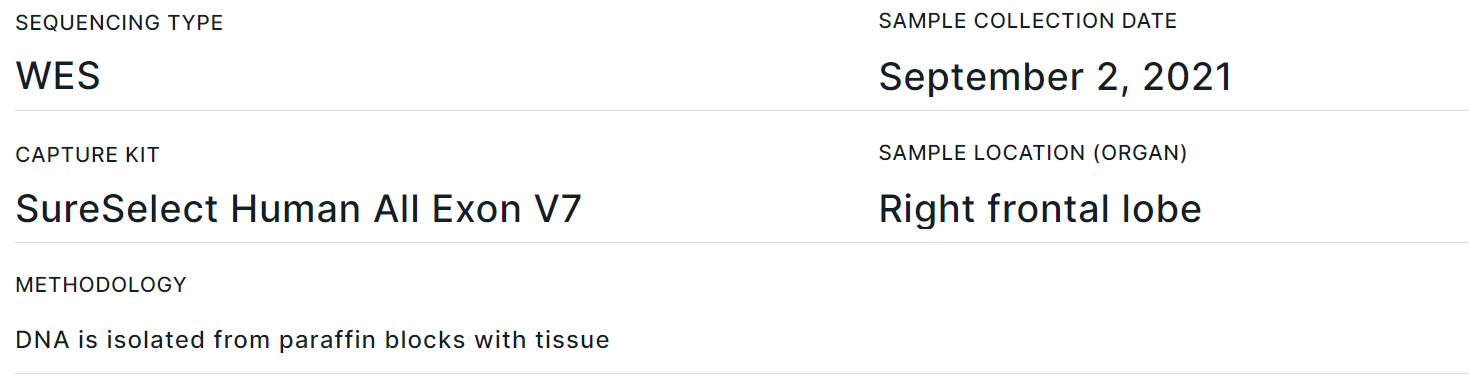
- Corresponds to "Sample info" template block.
- Contains basic data about the tumor sample: Sequencing type, Sample collection date, Capture Kit, Sample location (Organ), Methodology. You can also add fields with sample Short name, Sample type, Oncological disease, Onco stage, Onco stage details, Therapy line and Comments in the report template.
- Section fields can be filled in or edited on the report page, on the tumor sample page, or on "Sample" tab of the variant details page. Information is updated in the report automatically, regardless of where the data was updated.
Tier I: Variants of Strong Clinical Significance#
- Corresponds to "Glioma: Tier I variants" template block.
- Filled with data if somatic variants were discovered and successfully annotated in the tumor sample ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages).
- Contains onco-markers (somatic single nucleotide variants, indels, copy number variations and structural variations) of strong clinical significance associated with gliomas discovered in a tumor sample. Such onco-markers are very important for diagnostics and treatment of gliomas (Tier I).
Hereinafter, single nucleotide variants and indels are presented as following:#

- Gene is the common name of the gene in which the variant is located. If you click on a gene, you will see a window with all the gene transcripts:

You can find the description of the transcripts' table columns in the description of "Transcripts" section on the variant details panel.
- Position is a coordinate of the variant in the genome (chromosome + start position).
- Genetic variant is the nucleotide and amino acid substitutions using the HGVS notation. Nucleotide substitution: “c.” (coding; for a substitution in the coding sequence) or “n.” (non-coding; for a substitution in the non-coding sequence) prefix + genomic position of the substituted nucleotide + reference allele > alternative allele. Amino acid substitution: “p.” prefix (protein) + reference amino acid + amino acid position in protein + new amino acid resulting from the substitution.
- Transcript is the main transcript ID from the RefSeq database (NM_xxxxxx.x). If you click on a transcript, you will see the same window with all the gene transcripts, as when you click on the "Gene" field.
- Consequence is the effect of the variant on genes. A detailed description of the possible values can be found here.
- Allele frequency is an alternative allele frequency for the sample (in percentages).
- Coverage depth is a sequencing depth; the total number of reads of the sequence overlapping the variant position for the sample.
- External links are links to pages with variant information in dbSNP, ClinVar and COSMIC (if it was uploaded as a custom annotation).
- Links to the variant in embedded services:
is a module for visualization of variant on the genome,
is a variant details page in SNV Viewer ("Annotation" tab).
Hereinafter, structural and copy number variations presented as following:
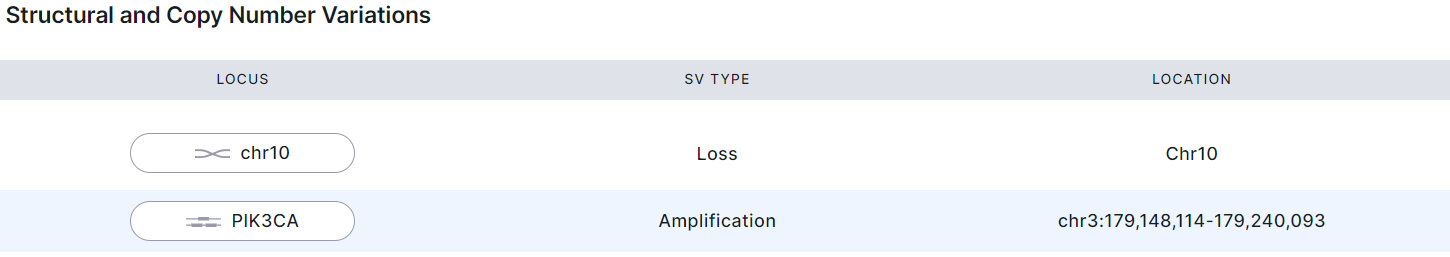
- Locus is the genome region affected by the variation (gene, chromosome, chromosome arm).
- SV type: deletion, duplication, inversion, translocation, amplification (for structural variations) and copy number gain/loss (for copy number variations).
- Location is the variation coordinates in the genome (chromosome (if the entire chromosome is affected); chromosome + arm; chromosome + start and final positions on it).
Tier II: Variants of Potential Clinical Significance#
- Corresponds to "Glioma: Tier II variants" template block.
- Filled with data if somatic variants were discovered and successfully annotated in the tumor sample ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages).
- Contains onco-markers (somatic single nucleotide variants, indels, copy number variations and structural variations) of potential clinical significance discovered in a tumor sample. These variations are likely associated with gliomas, but this association is less well studied compared to Tier I variations.
Immunotherapy Markers#
- Corresponds to "Immunotherapy markers" template block.
- Filled with data if tumor sample was uploaded in FASTQ or BAM format and the somatic variants were discovered and successfully annotated in this sample ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages).
- Contains a calculated tumor mutation burden (TMB) level, which is the average number of non-inherited somatic mutations per million bases (Mb). High TMB (more than 19 mutations per 1 Mb) may predict response to immunotherapy, and low TMB (less than 6 mutations per 1 Mb) may predict no susceptibility to immunotherapy.

Suggested Diagnosis#
- Corresponds to "Glioma: Suggested diagnosis" template block.
- Filled with data if tumor sample was uploaded in FASTQ or BAM format and the somatic variants were discovered and successfully annotated ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages) and the copy number variations were successfully discovered ("Copy number variations discovery" stage) in this sample.
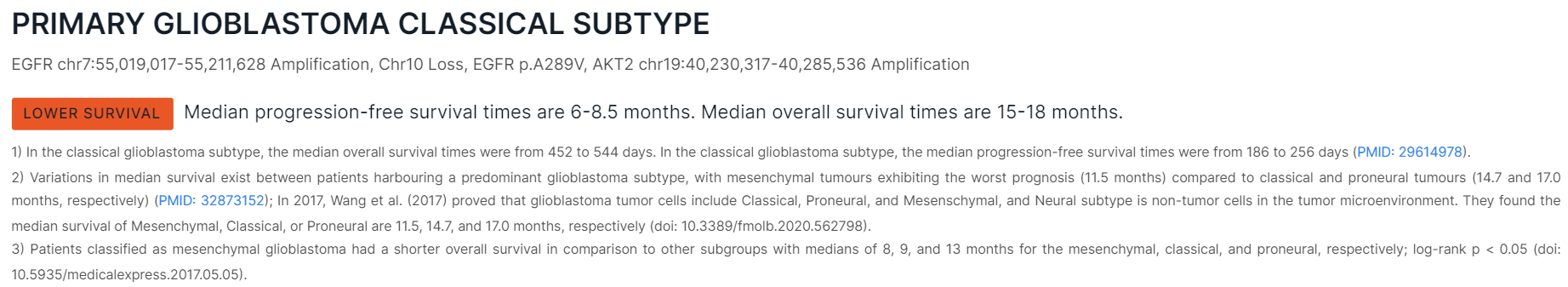
- Contains a suggested diagnosis based on Tier I and Tier II mutations discovered in the sample and a knowledge base containing more than 170 markers (single nucleotide variants, indels, structural variations, copy number variations) associated with glioma. Suggestions are made under the assumption that the tumor sample DNA has been extracted from glioma. In case of uncertainty, up to three most probable diagnosis suggestions are proposed.
- For a specific diagnosis, there are a rationale as a list of Tier I and Tier II markers, the predicted survival times with excerpts from the publications from which the prediction was made, and links to publications.
caution
All interpretations are based on published research and are provided for informational purposes only. They are not intended to be a substitute for professional medical advice, diagnosis, or treatment.
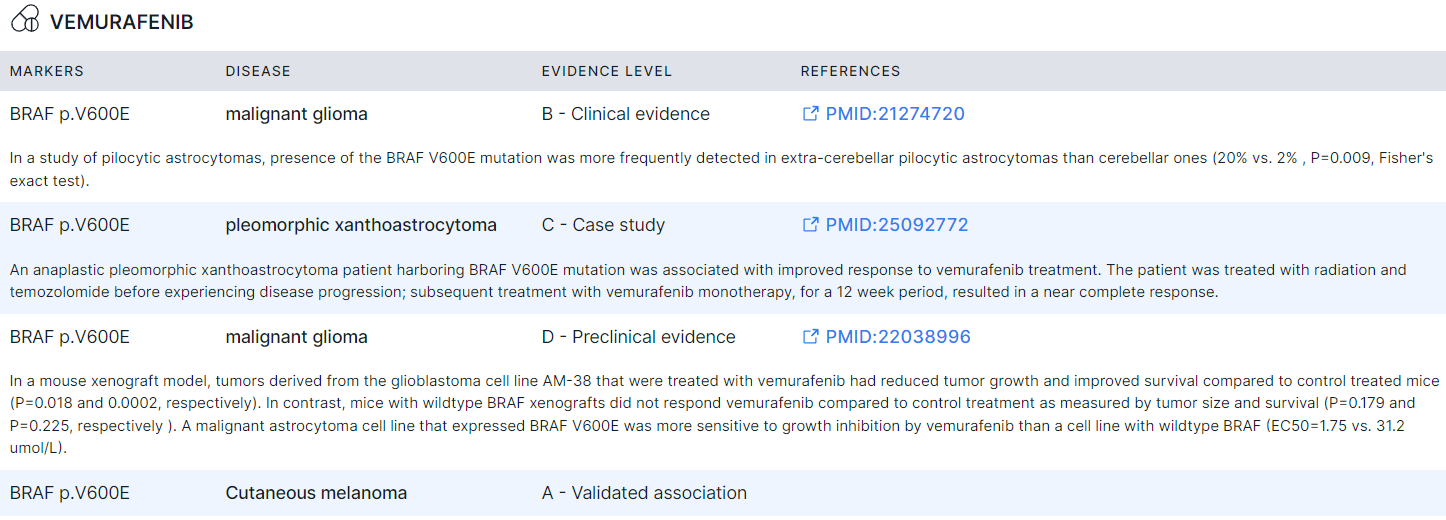
Suggested Drugs#
- Corresponds to "Glioma: Suggested drugs" template block.
- Filled with data if the somatic variants were discovered and successfully annotated in the tumor sample ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages).
- Contains tumor-targeting drugs that are potentially effective and ineffective against glioma, selected in CIVIC and CGI databases based on the biomarkers discovered in the sample.
- For each drug there is a table with the following columns:
- Markers are biomarkers (single nucleotide variant, indel, structural variation or copy number variation) of Tier I or Tier II that were discovered in the sample and which correspond to the drug.
- Disease for treatment which the drug is approved (if the marker was discovered).
- Evidence level describes the reliability of the study supporting the evidence:
- A (validated association): a good research-based evidence to support association;
- B (clinical evidence): a fair research-based evidence to support association, confirmed by clinical trials or other primary data;
- C (case study): the association is shown in individual case reports from clinical journals;
- D (preclinical evidence): the association supported by in vivo and in vitro models.
- References: a link to a publication that confirms the drug as tumor-targeting if this marker
is discovered.
For some markers, an excerpt from a publication with proof of association is provided.
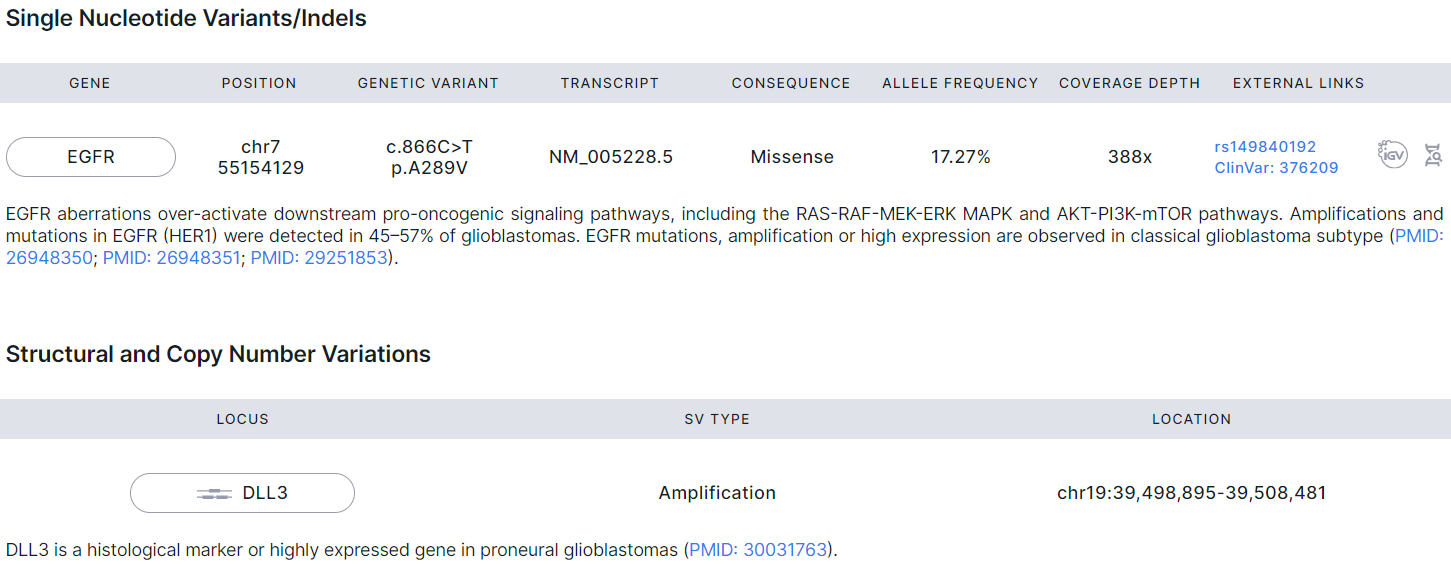
Mutations Details#
- Corresponds to "Glioma: Mutations details" template block.
- Includes Tier I and Tier II mutations with excerpts from scientific publications confirming the association of the mutation with gliomas with links to the original publications.
Tier III: Mutations in Other Cancer-Panel Genes#
- Corresponds to "Glioma: Tier III variants" template block.
- Filled with data if the somatic variants were discovered and successfully annotated in the tumor sample ("Somatic SNVs/Indels discovery" and "Somatic SNVs/Indels annotation" stages).
- Contains onco-markers (somatic single nucleotide variants, indels, copy number variations and structural variations) of high gene product impact detected in genes associated with cancers other than glioma (Tier III).
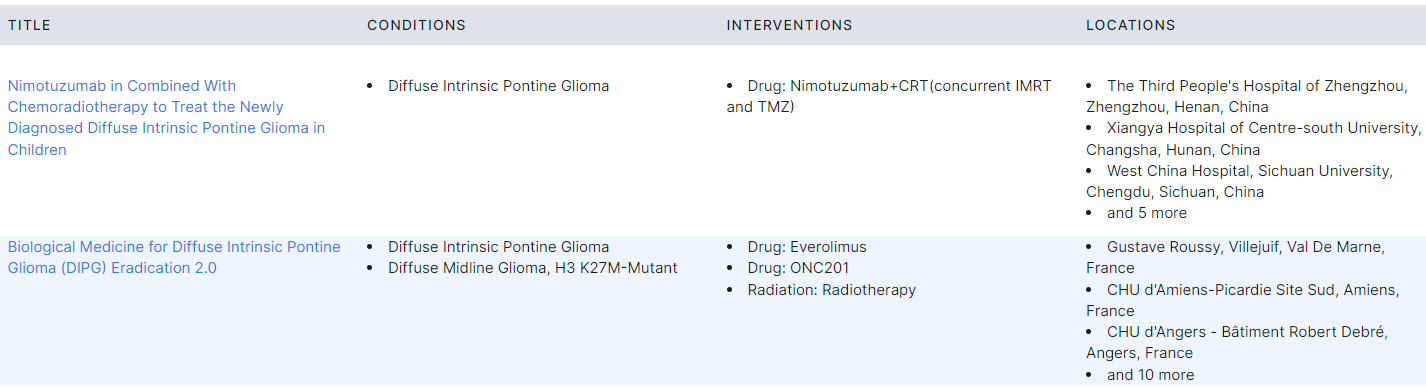
Clinical Trials#
- Corresponds to "Clinical trials" template block.
- Lists ongoing clinical trials of various interventions (drugs, devices, procedures, etc.) involving patients with a specific phenotype selected in the template block (in this case, glioma) from ClinicalTrials database.
- The table with clinical trials has the following columns:
- Title of the study with a link to a page with detailed information about it in ClinicalTrials database.
- Conditions is the disease, disorder, syndrome, illness, or injury being studied. It may also include other health-related issues, such as lifespan, quality of life, and health risks.
- Interventions are processes or actions that are the focus of a clinical study. Interventions include drugs, medical devices, procedures, vaccines, and other products that are either investigational or already available. Interventions can also include noninvasive approaches, such as education or modifying diet and exercise.
- Locations are locations of the studies (institution, city, country).
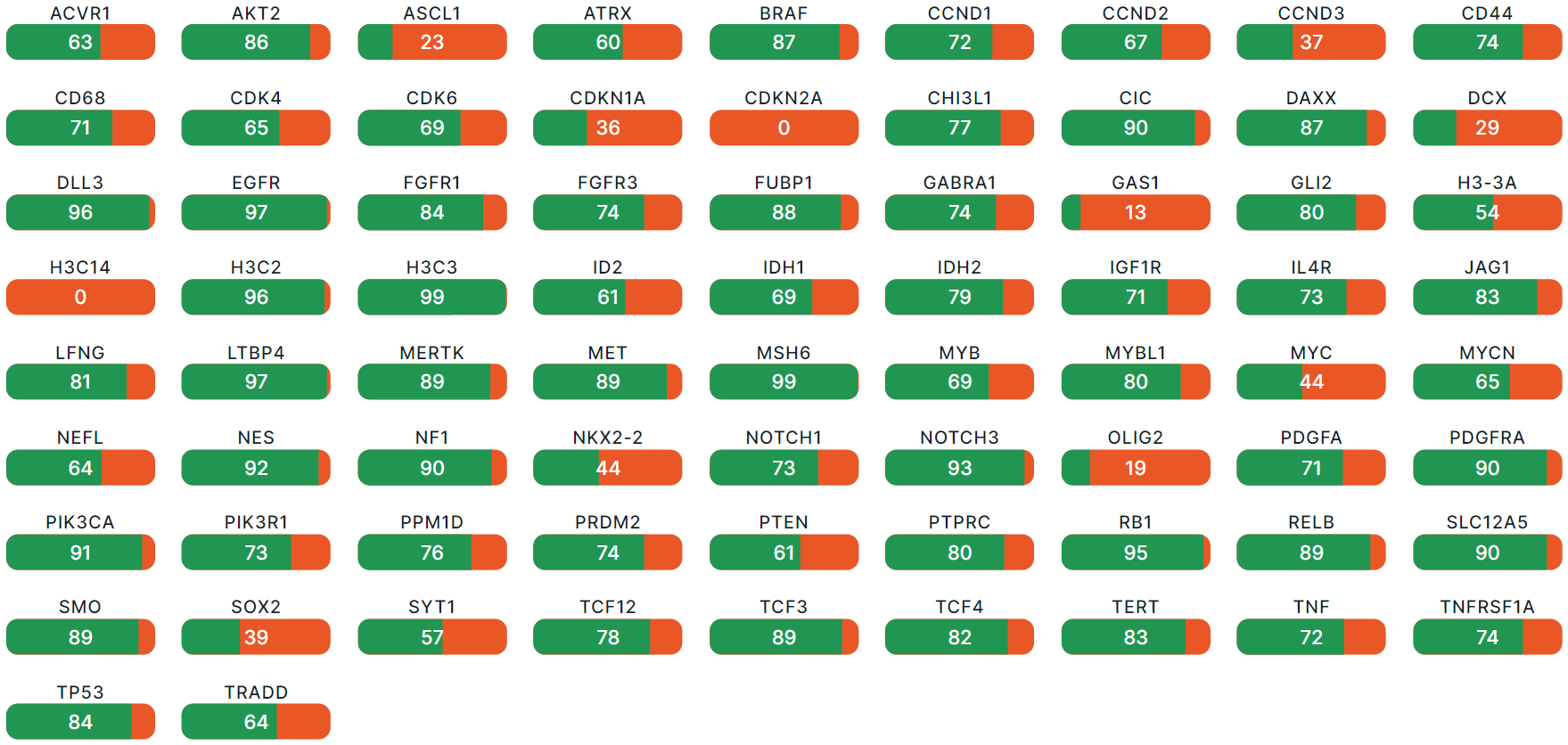
Glioma Genes Panel Details#
- Corresponds to "Glioma: Glioma genes panel details" template block.
- Filled with data if tumor sample was uploaded in FASTQ or BAM format.
- Contains information on coverage (fraction of the exonic part of the longest transcript covered by at least 50 sequenced reads (50X)) of glioma-specific genes with strong or potential clinical significance related to glioma, discovered in the tumor sample. Shows an assessment of the completeness and quality of the source data for this report. The higher the percent of coverage, the more completely a transcript is sequenced.
2. Germline Variants Report#
- The section contains information obtained from the analysis of a non-tumor (control) sample. It is support section containing germline variants that are pathogenic and/or associated with gliomas.
Control Sample Info#
- Corresponds to "Control (normal) sample info" template block.
- Filled with data if the analyzed sample set includes a non-tumor (control) sample in addition to the tumor sample.
- Contains basic data about the control sample: Sequencing type, Sample collection date, Capture Kit, Sample location (Organ), Methodology. You can also add fields with sample Short name, Sample type, Oncological disease, Onco stage, Onco stage details, Therapy line and Comments in the report template.
- Section fields can be filled in or edited on the report page, on the non-tumor sample page, or on "Sample" tab of the variant details page. Information is updated in the report automatically, regardless of where the data was updated.
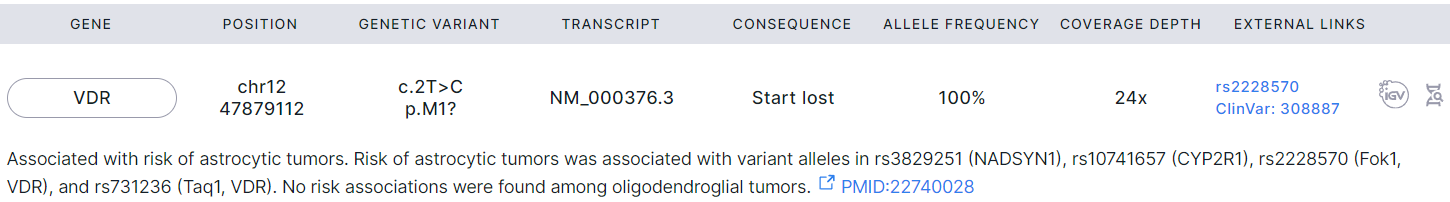
Pathogenic or Likely Pathogenic for Gliomas#
- Corresponds to "Glioma: Germline pathogenic for gliomas" template block.
- Filled with data if the germline variants were discovered and successfully annotated in a non-tumor (control) sample or a single tumor sample ("Germline SNVs/Indels discovery" and "Germline SNVs/Indels annotation" stages).
- Contains single nucleotide germline variants, discovered in the control sample and associated with gliomas according to published research. These variants meet at least one of the following conditions:
- Marked as pathogenic in ClinVar;
- Marked as likely pathogenic in ClinVar;
- Have high gene product impact;
- Have moderate gene product impact and frequency less than 10% in population;
- Rare (have frequency less than 5% in population).
- The variant interpretation text may be given below the variant row if it was added as described here.
- Next, there are excerpts from scientific publications for each variant confirming its association with gliomas with links to the publications.
ACMG secondary findings#
- Corresponds to "SNVs/Indels in ACMG SF genes" template block.
- Filled with data if the samples were uploaded in FASTQ or BAM formats or as VCF files (each with a single sample) and if germline variants were discovered and successfully annotated in a non-tumor (control) sample or a single tumor sample ("Germline SNVs/Indels discovery" and "Germline SNVs/Indels annotation" stages).
- Includes the variants discovered in the sample, which are located in the genes from the Secondary Findings (SF) list of American College of Medical Genetics and Genomics (ACMG). For these genes, specific mutations are known to be causative of disorders with defined phenotypes that are clinically actionable by an accepted intervention. The ACMG recommends that variants detected in any of these genes should be reported as they are of medical relevance and could be used in the future to inform clinical treatment.
- The variants are divided into two lists according to the certain conditions, which are described in the corresponding section.
caution
Annotating variants with ClinVar to generate ACMG secondary findings report has the limitations described here.
Pathogenic or Likely Pathogenic for Any Other Genes/Diseases#
- Corresponds to "Glioma: Germline pathogenic for any other diseases" template block.
- Filled with data if the germline variants were discovered and successfully annotated in a non-tumor (control) sample or a single tumor sample ("Germline SNVs/Indels discovery" and "Germline SNVs/Indels annotation" stages).
- Contains single nucleotide germline variants discovered in the sample, marked as pathogenic or likely pathogenic in ClinVar and associated with diseases other than gliomas.
- The variant interpretation text may be given below the variant row if it was added as described here.
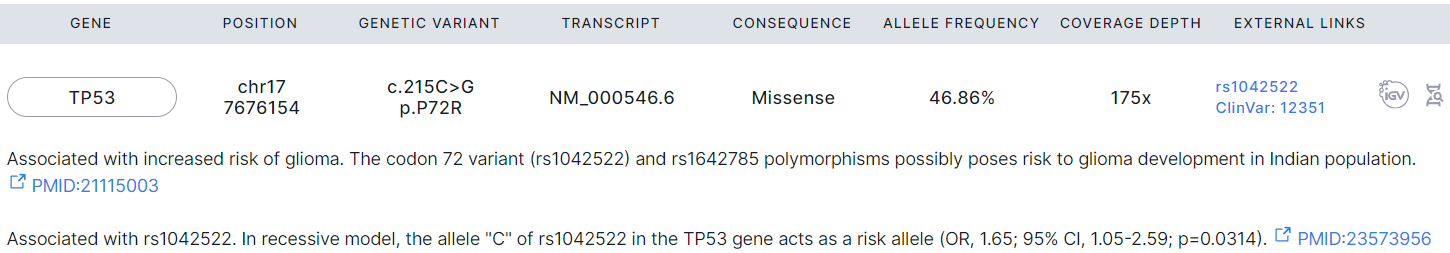
Glioma Associated#
- Corresponds to "Glioma: Germline glioma associated" template block.
- Filled with data if the germline variants were discovered and successfully annotated in a non-tumor (control) sample or a single tumor sample ("Germline SNVs/Indels discovery" and "Germline SNVs/Indels annotation" stages).
- Contains single nucleotide germline variants discovered in the sample, associated with gliomas according to published research, but not included in "Pathogenic or likely pathogenic for gliomas" section.
- The variant interpretation text may be given below the variant row if it was added as described here.
- Next, there are excerpts from scientific publications for each variant confirming its association with gliomas with links to the publications.
3. Reference List#
- Corresponds to "Glioma: Literature" template block.
- Contains links to articles, publications, and books related to glioma research that were used in compiling the report.
4. Technical Limitations#
- Corresponds to "Disclaimer" template block.
- Contains a message about the limitations of the use of the report (the text can be edited in the template).
Report export#
Glioma report can be downloaded in PDF format. To do this, click on
the button in the upper right corner of the
report page.