Filtering of Variations
The needed copy number variations can be found by filtering. Filtering options are located above the table:

note
Table filtering is saved when you exit the CNV Viewer page.
1. Log2FC thresholds#
Variation filtering by log2FC thresholds - the logarithmic ratio of the detected copy number to the normal copy number. The available threshold values for this filter depend on the sample's chromosomal sex determined by CNVkit from the relative copy number of the autosomes and sex chromosomes.
If the chromosomal sex is determined as male (XY) or if sex cannot be determined, the available threshold values will be:
- Del: ≤-1, Dup: ≥0.58 (chrX: ≥1, chrY: ≥1) (XY)
- Del: ≤-0.9 (chrX: ≤-3.8, chrY: ≤-3.8), Dup: ≥0.55 (chrX: ≥0.94, chrY: ≥0.94) (XY)
- Del: ≤-0.8 (chrX: ≤-2.74, chrY: ≤-2.74), Dup: ≥0.51 (chrX: ≥0.88, chrY: ≥0.88) (XY)
- Del: ≤-0.7 (chrX: ≤-2.11, chrY: ≤-2.11), Dup: ≥0.46 (chrX: ≥0.82, chrY: ≥0.82) (XY) - default value
- Del: ≤-0.6 (chrX: ≤-1.64, chrY: ≤-1.64), Dup: ≥0.42 (chrX: ≥0.74, chrY: ≥0.74) (XY)
- Del: ≤-0.5 (chrX: ≤-1.27, chrY: ≤-1.27), Dup: ≥0.37 (chrX: ≥0.66, chrY: ≥0.66) (XY)
- Del: ≤-0.4 (chrX: ≤-0.95, chrY: ≤-0.95), Dup: ≥0.31 (chrX: ≥0.56, chrY: ≥0.56) (XY)
- Del: ≤-0.3 (chrX: ≤-0.67, chrY: ≤-0.67), Dup: ≥0.24 (chrX: ≥0.45, chrY: ≥0.45) (XY)
If the chromosomal sex is determined as female (XX), the available threshold values will be:
- Del: ≤-1 (chrX: ≤0), Dup: ≥0.58 (chrX: ≥1.58, chrY: ≥0) (XX)
- Del: ≤-0.9 (chrX: ≤0.1), Dup: ≥0.55 (chrX: ≥1.55, chrY: ≥-0.1) (XX)
- Del: ≤-0.8 (chrX: ≤0.2), Dup: ≥0.51 (chrX: ≥1.51, chrY: ≥-0.23) (XX)
- Del: ≤-0.7 (chrX: ≤0.3), Dup: ≥0.46 (chrX: ≥1.46, chrY: ≥-0.37) (XX) - default value
- Del: ≤-0.6 (chrX: ≤0.4), Dup: ≥0.42 (chrX: ≥1.42, chrY: ≥-0.55) (XX)
- Del: ≤-0.5 (chrX: ≤0.5), Dup: ≥0.37 (chrX: ≥1.37, chrY: ≥-0.77) (XX)
- Del: ≤-0.4 (chrX: ≤0.6), Dup: ≥0.31 (chrX: ≥1.31, chrY: ≥-1.04) (XX)
- Del: ≤-0.3 (chrX: ≤0.7), Dup: ≥0.24 (chrX: ≥1.24, chrY: ≥-1.41) (XX)
Filtering algorithm::
- Click on the filter window;
- Select the needed filtering option from the drop-down list, or type the option you are looking for in the search box and select from the found.
Done! Filter applied. The selected filter can be seen in the window:
When all filters are reset, the log2FC threshold filter reverts to its default value. This value can only be changed or cleared within the filter itself (see below for details).
Let’s take default value for XY (Del: ≤-0.7 (chrX: ≤-2.11, chrY: ≤-2.11), Dup: ≥0.46 (chrX: ≥0.82, chrY: ≥0.82)) as an example to explain how these filtering conditions work:
- Copy number variations on autosomes with log2FC ≤ -0.7 are considered deletions. -0.7 is the default log2FC threshold, below which only one autosome is called instead of the normal two.
- Copy number variations on the X and Y chromosomes with log2FC ≤ -2.11 are considered deletions.
- Copy number variations on autosomes with log2FC ≥ 0.46 are considered duplications. The threshold for duplication on an autosome is calculated using the following formula: log2FC = log2((2+D)/2), where D is the “number” of copy number that is “lost” at log2FC = -0.7, and 2 is the number of sister chromatids in the autosome.
- Copy number variations on the X and Y chromosomes with log2FC ≥ 0.82 are considered duplications.
- Variations that do not satisfy the thresholds, i.e. variations on autosomes with -0.7 < log2FC < 0.46 and variations on sex chromosomes with -2.11 < log2FC < 0.82 do not pass filtering, i.e. are excluded from the CNV Viewer table.
Calculation of log2FC thresholds for deletions and duplications on autosomes and sex chromosomes#
The threshold of calling a single copy instead of two on autosomes is calculated as
follows: T21 = log2(C/2), where C is the number of called
copies.
Delta of copy number: D = 2 – C = 2 – 2 × 2T21.
Then the corresponding threshold of calling three copies instead of two on autosomes is calculated as
follows: T23 = log2((2+D)/2) = log2(2-2T21).
Threshold of calling duplications on autosomes: hi = T23.
To calculate the threshold of calling three copies instead of two on the X chromosome, we normalize to XY genotype by dividing it by copy number of 1: T23X= log2((2+D)/1) = log2(2+2-2×2T21) = 1 + log2(2-2T21) = 1 + T23.
Calculating the threshold of calling duplications on the X chromosome:
- if there is no Y chromosome: Xhi = 1 + T23;
- if there is Y chromosome: Xhi = log2(3-2×2T21).
The threshold of calling one copy instead of two on the X chromosome: T21X = log2((2-D)/1) = log2(2-2+2×2T21) = 1 + T21.
Threshold of calling no copies instead of one on the X chromosome: T10X = log2((1-D)/1) = log2(1-2+2×2T21) = log2(-1+2×2T21).
Calculating the threshold of calling deletions on the X chromosome:
- if there is no Y chromosome: Xlo = 1 + T21;
- if there is Y chromosome: Xlo = log2(-1+2×2T21).
The threshold of calling one copy instead of none on the Y chromosome in the case of genotype XX: T01 = log2((0+D)/1) = log2(2-2×2T21) = log2(2×(1-2T21)) = 1 + log2(1-2T21).
Calculating the threshold of calling duplication Yhi and deletions Ylo on the Y chromosome:
- if there is no Y chromosome: Yhi = 1 + log2(1-2T21)); Ylo = -∞;
- if there is Y chromosome: Yhi = Xhi; Ylo = Xlo.
2. Region chr:start-end#
Filter variations by position. To filter, enter the position of the needed genomic interval, i.e. the corresponding chromosome and the start and end positions. Possible options of writing a position:
chr1:161,190,824-161,193,921chr1:161190824-161193921chr1:161190824_1611939211:161190824-161193921
You can also filter by more than one interval at once. For example,
by chr1:161190824-161193921, chrX:151110769-151113866.
The filtering result will be rows with variations whose positions intersect with the entered interval.
3. Gene panel#
Filtering variations by localization in certain genes from the panel. You can add a gene panel on "Gene Panels" page or import a panel from the Library of the most common clinical panels. To import, click on "Import from Library":

Select the required panels by ticking them in the window that opens:
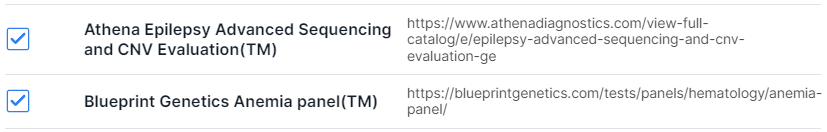
and click on to add panel.
4. Gene#
Filtering variations by localization in the gene transcript. The transcript is given in brackets next to the official gene name. You can filter variations by several genes at once. To do this, tick the required genes and click outside the value area:
If several transcripts are known for a gene, the list presents all of them:
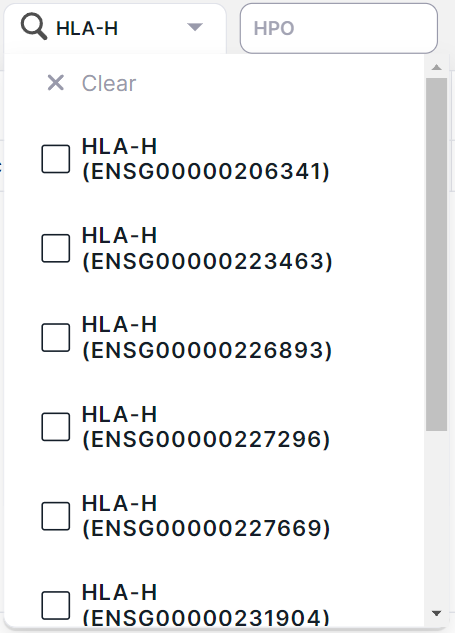
5. HPO#
Filtering variations by localization in genes associated with terms from Human Phenotype Ontology (HPO). Click on the HPO filter container to open the phenotype search form. The form presents the main groups of phenotypes in the drop-down list:
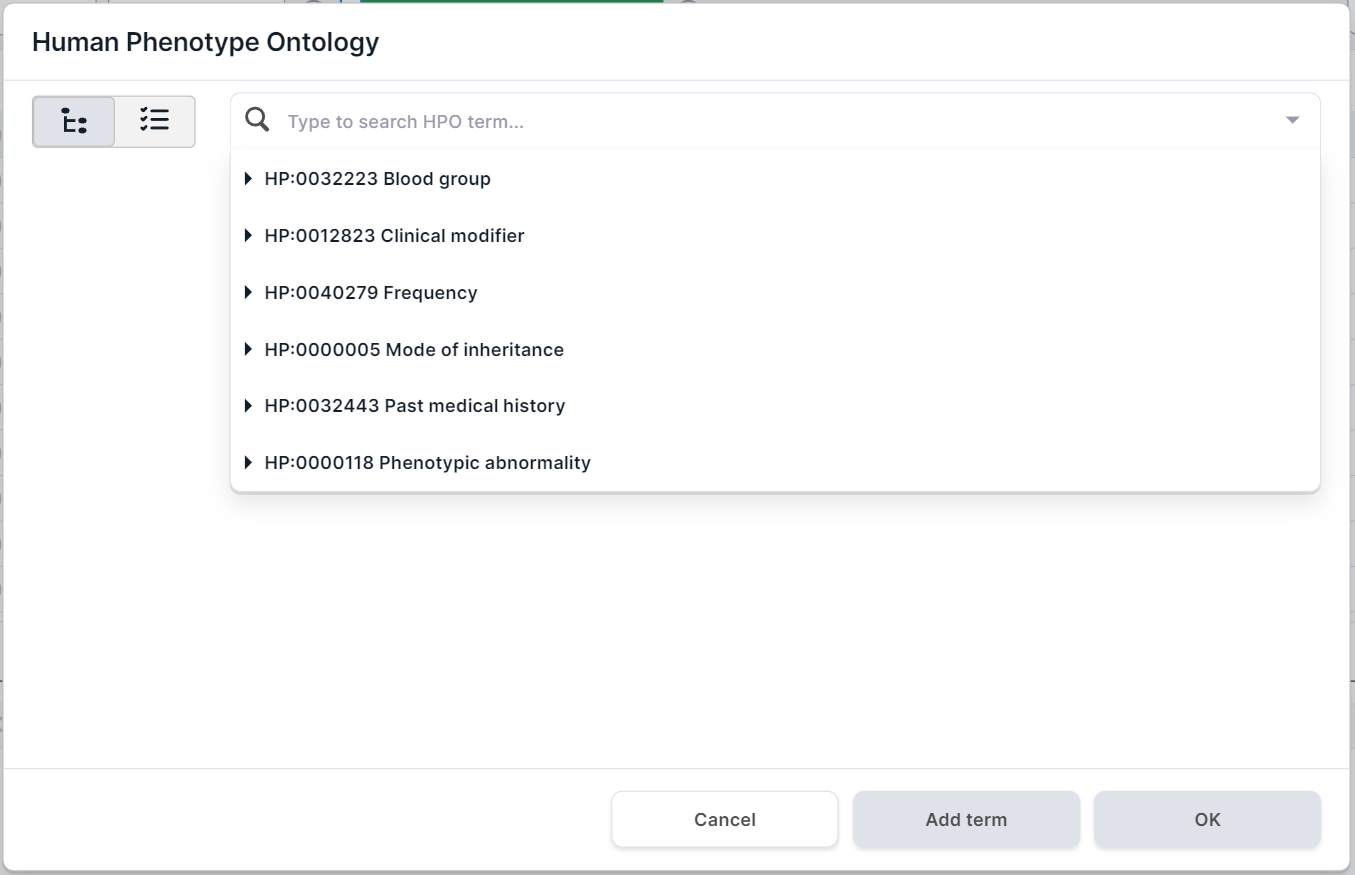
Ways to search and select a term for filtering:
- Search for a term in the drop-down list. You can interact with term groups
using
and
to expand and collapse the list of group phenotypes, respectively:
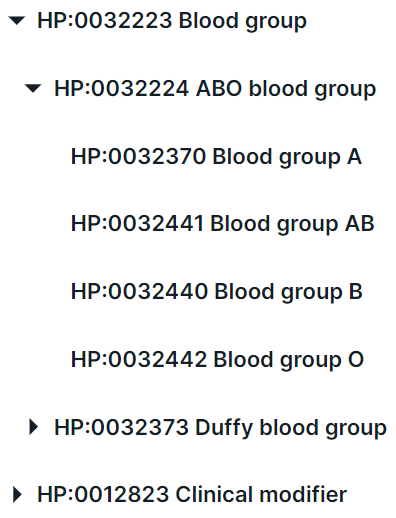
In addition, you can find a phenotype by searching for the name or ID from HPO. For example, when searching for the disease “glioma”, several terms are found:
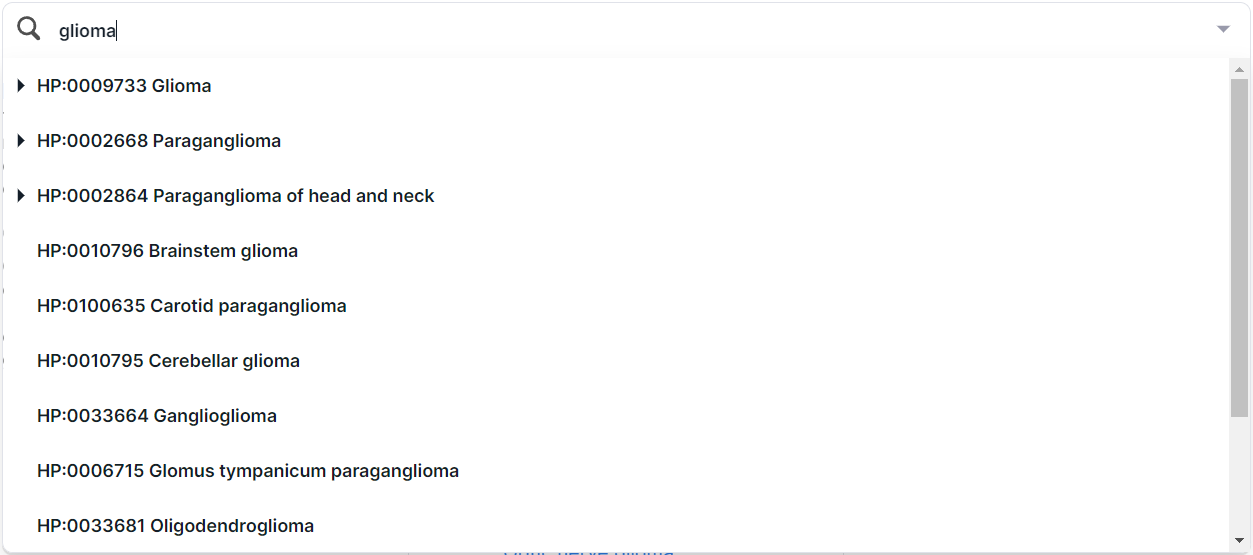
To select a term to filter, click on its row. You can choose both the whole group and a specific phenotype. By selecting one of the terms (e.g. "Glioma"), you will see this disease card:
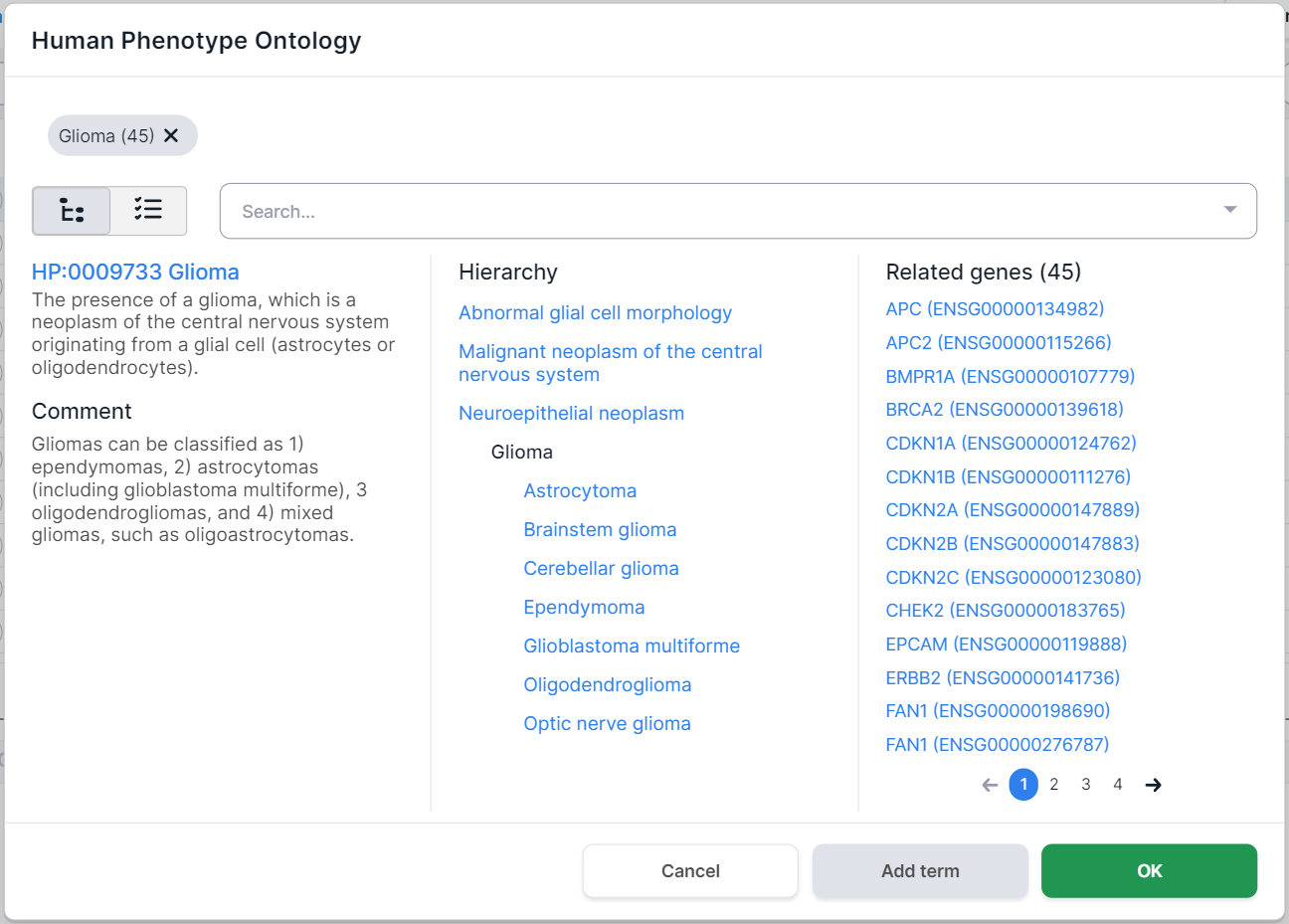
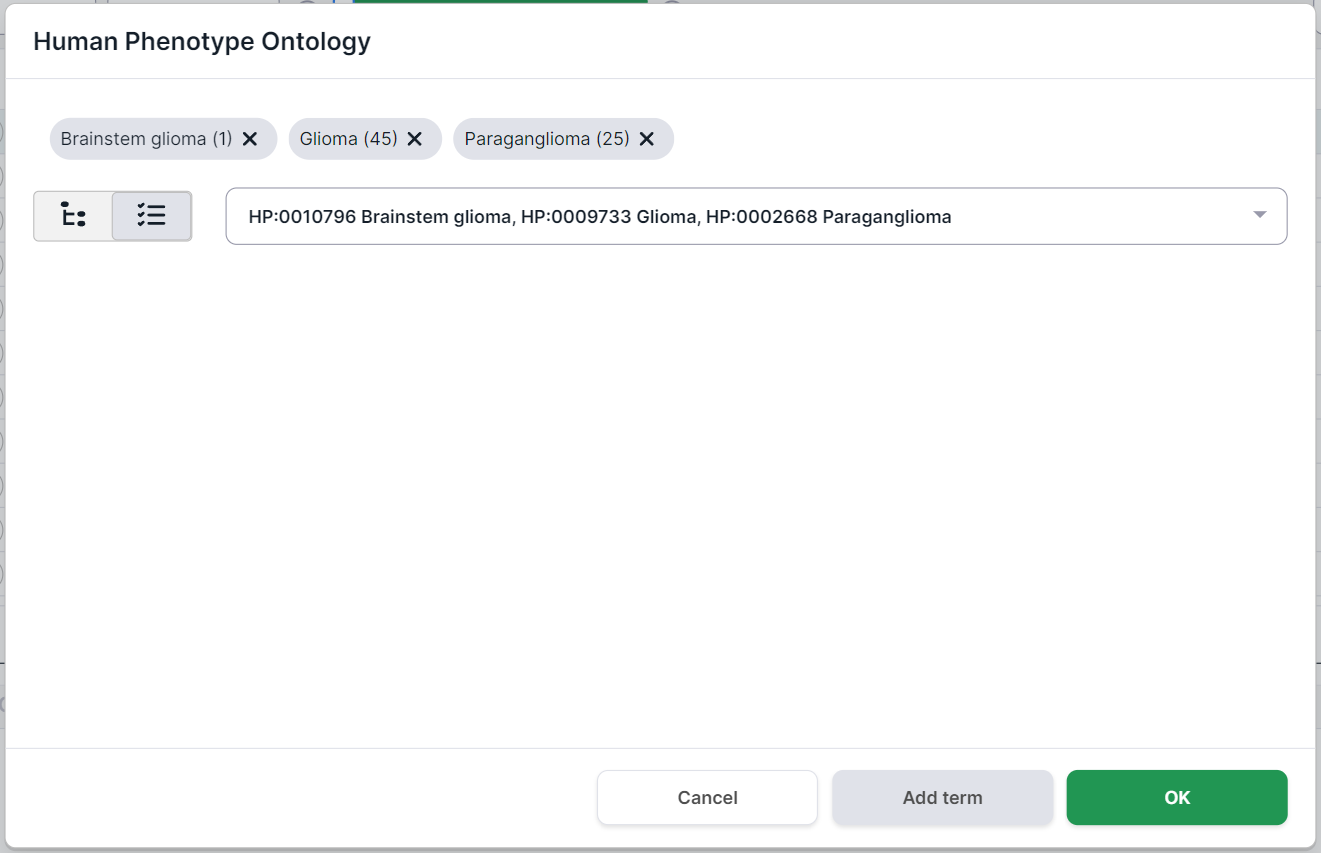
The phenotypes selected for filtering are marked above the disease card. Number in parentheses
indicates the number of genes associated with the phenotype. By clicking on a term, you will see
the phenotype card. You can remove the term by clicking on
the cross :

On the left side of the phenotype card, there are its definition and comments. In the middle, there is the hierarchy of the disease: the terms above the selected Glioma disease are its ancestor phenotypes (i.e. the phenotype groups that include Glioma), and the terms below Glioma are its child phenotypes (i.e. the diseases that are part of the Glioma group). On the right, there are the genes associated with this disease (in this case, 45 genes were found).
Search for a term by hierarchy in the phenotype card. By clicking on the name of the ancestor or child phenotype, you will see this phenotype page. If the phenotype has not yet been selected for filtering and genes associated with it have been found, then you can select it for filtering variations by clicking on
.
Search for a term in multiselect mode. Switch the search mode by clicking on
. You will see a list of main groups of phenotypes with checkboxes. You can find a phenotype by searching for the name or ID from HPO. For example, when searching for the disease “glioma”, several terms are found:
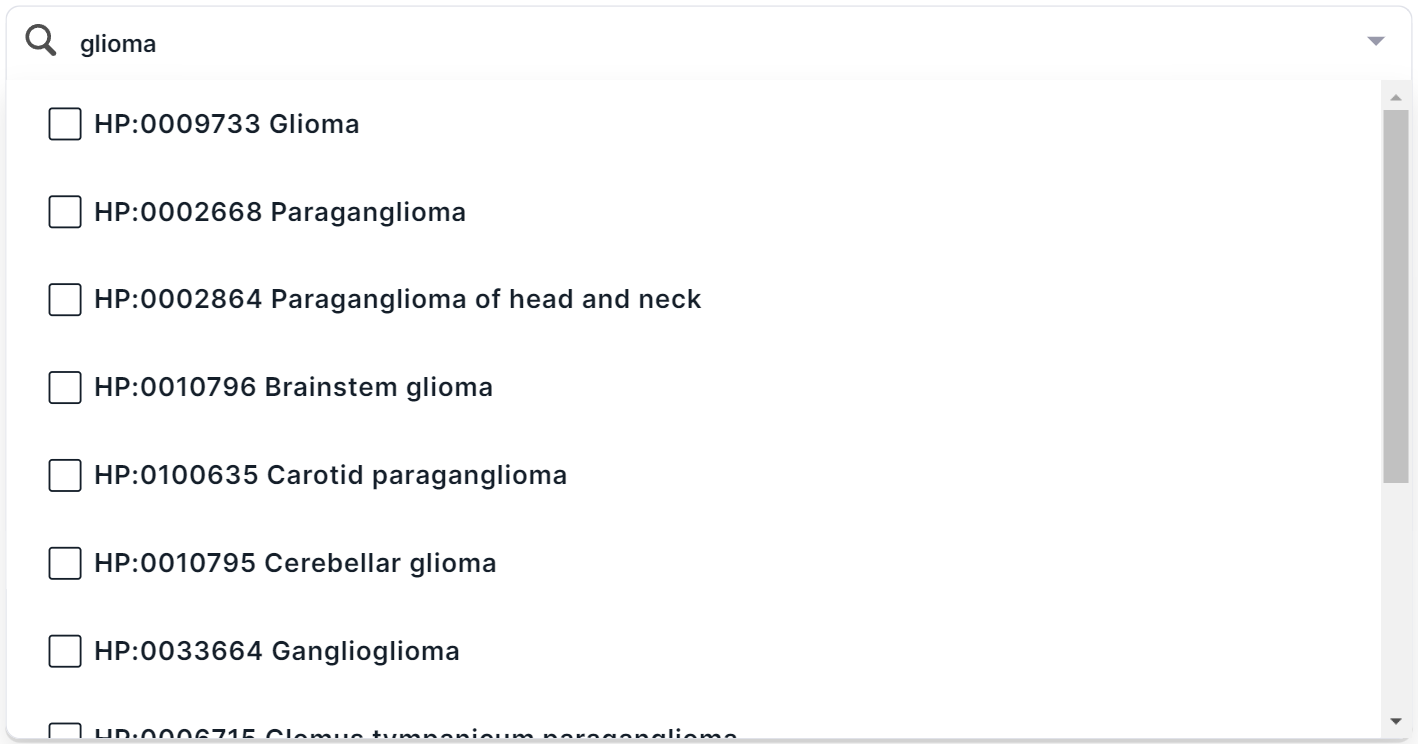
In multiselect mode, you can select several terms at once in the drop-down list. To select a term to filter, click on its line:
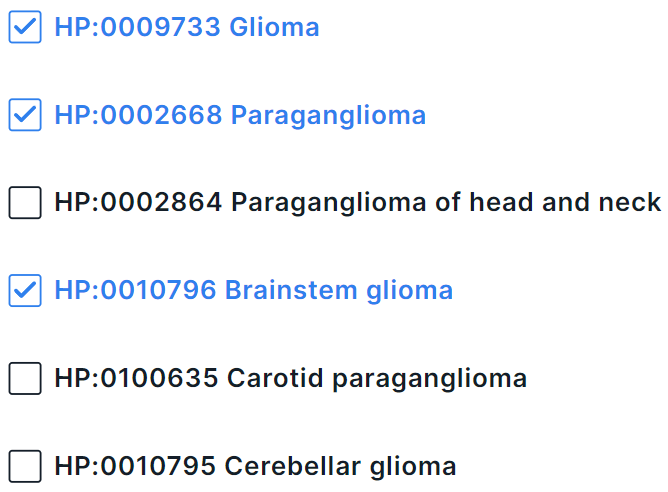
To complete adding terms for filtering, click outside the drop-down box within the phenotype search form.
At the top of the form, you will see the phenotypes selected for filtering. Number in parentheses
indicates the number of genes associated with the phenotype. By clicking on a term, you will see
the phenotype card. You can remove the term by clicking on
the cross .
To filter variations by genes associated with the selected phenotypes,
click on .
You can see the selected phenotypes in the tooltip by hovering over the HPO filter field:

To cancel filtering by all phenotypes, hover over the filter window and click on the cross:
or click on the field and edit your selection in the phenotype search form.
6. Type#
Filter variations by the copy number variation type: LOSS - deletion, GAIN - duplication.
7. CDS Overlap#
Filter variations by overlapping with CoDing Sequence (CDS):
- Overlapped CDS - the variation affects the CDS (percentage of overlapped CDS is greater than 0);
- Not overlapped CDS - the variation does not affect the CDS (percentage of overlapped CDS equals 0).
8. ACMG Class#
Filter variations by pathogenicity assigned according to the ranking score defined by AnnotSV:
- Pathogenic;
- Likely pathogenic;
- Uncertain significance conflict - conflicting interpretations of pathogenicity;
- Uncertain significance;
- Likely benign;
- Benign;
- Undefined.
You can filter variations by several pathogenicity values at once. To do this, tick the required values and click outside the value area.
9. Cohort 180 AF#
Filter variations by frequency in a cohort of 188 normal samples. AF values are only available for germline CNVs discovered in WGS or low-pass WGS samples normalized to the "BGI WGS" reference samples panel with bin size 3000 or 5000 or the "BGI WGS v2" reference samples panel with bin size 3000. The filter is only displayed in the CNV Viewer with copy number variations discovered in the bins.
Possible values:
- <0.05 or empty;
- <0.02 or empty;
- <0.01 or empty;
- <0.005 or empty;
- <0.0001 or empty.
Reset Filters#
To reset all applied filters, click on .
Only filtering by default log2FC threshold
value - Del: ≤-0.7 (chrX: ≤-2.11, chrY: ≤-2.11), Dup: ≥0.46 (chrX: ≥0.82, chrY: ≥0.82) will be preserved.
To clear a separate filter:
- Click on the filter window and then on
.
- To clear filtering by region, hover over the filter window and click on the cross:

- For a filter by gene, you can remove one of the selected filtering options. To do this, click on the filter window and uncheck the option.
- To change the selection of HPO phenotypes by which variations are filtered, click on the HPO filter window and edit your selection in the phenotype search form. To clear filtering by all selected phenotypes, hover over the filter window and click on the cross: