Check quality and cleanup
Quality control is an important step in sequencing data analysis. It is initiated after the "Upload, identify and verify" stage has successfully completed. First, the reads in the sample file(s) are checked to ensure that they meet the quality criteria. Then, if cleanup is enabled and the sample does not meet the quality criteria, cleanup is initiated for the sample. Cleanup may involve filtering out reads that do not meet the quality criteria and/or trimming adapter sequences, primers, poly(A) tails, and other types of sequences from the reads. Cleanup can significantly improve the quality of mapping and variant discovery.
If a single read sequencing sample is analyzed, quality control will be run for only one file and the corresponding workflow stage will be called "Check quality and cleanup". If a paired read sequencing sample is analyzed, then quality control will be run for two paired files and there will be two corresponding workflow stages: "Check quality and cleanup primary" and "Check quality and cleanup mate".
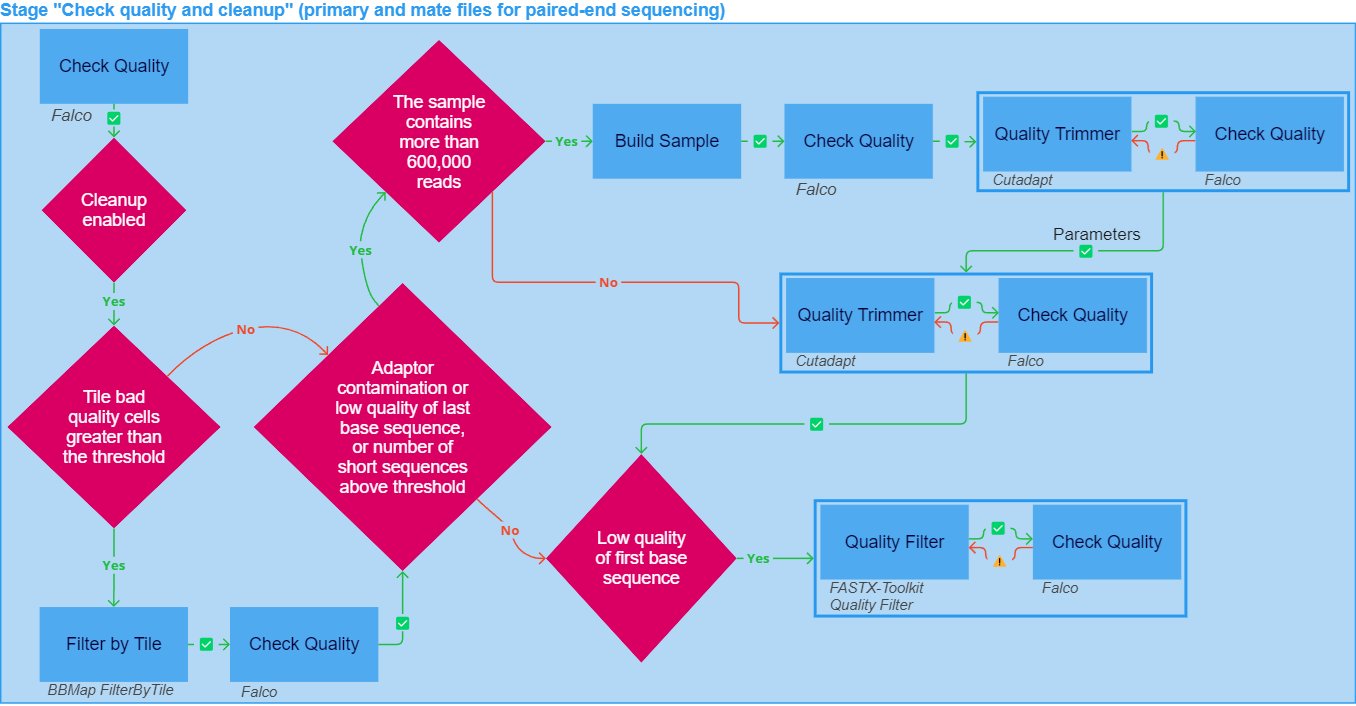
The "Check quality and cleanup" stage of sample analysis may include the following tasks:
- Check Quality of reads in the sample file(s) using Falco, which is on average three times faster than the equivalent FastQC1 tool. In addition, the quality format of the sample file is determined. The quality format is written as Q+number (e.g. "Q33"), where Q is the Phred quality score, a measure of the quality of the identification of the nucleobases generated by automated DNA sequencing, and 33 is the ASCII base. The Q33 format is newer and is used by Sanger and Illumina 1.8, while the Q64 format is older and is used by earlier Illuminas.
A detailed report with visualized results of the quality check of the sample reads for each quality control metric can be opened in the "Result files" section in the "Check Quality" task details ("Open Quality Report HTML"). There you can also download the same report in text form ("Download Quality Report Data TXT"). The results for each quality control metric are provided below, in the "Metrics" section in the "Check Quality" task details.

For each metric, the following are provided:
- metric name;
- description of the result of checking the compliance of a certain indicator of sample reads and the threshold of this metric: the indicator value for the sample (e.g. Total sequences, i.e. the total number of sequences in the file) and the used threshold of the metric, at which reads in the sample are considered high-quality (can be changed in the parameters) are provided;
- result of quality check by metric:
if the sample satisfies the threshold of the metric, or
if the sample does not satisfy it.
The sequence quality control metrics#
| Metric | The metric threshold value at which reads in a sample are considered high-quality (this is the default value, can be changed in parameters) | Consequences if sample does not meet metric threshold |
| Total sequences | The minimal meaningful number of reads in a file is 10,000. | Sample analysis is interrupted. |
| Length distribution | The file contains no more than 25% short reads (i.e. reads with length ≤ 20 bp). | The "Quality Trimmer" task: filtering short reads. |
| Tiles sequence quality | The file contains no more than 10 faulty tiles (i.e. tiles that exceed mean quality across all tiles by 7). | The "Filter by Tile" task: tiles that produced low quality reads are excluded from the analysis. |
| First base sequence quality | The quality of the three start positions in a read is not less than 20. | The "Quality Filter" task: filtering low quality reads. |
| Middle base sequence quality | The quality of middle positions in a read is not less than 20. | The "Quality Trimmer" or "Quality Filter" task. |
| Last base sequence quality | The quality of the three end positions in a read is not less than 20. | The "Quality Trimmer" task. |
| Overrepresented sequences | The file contains no more than 1% overrepresented sequences (i.e. sequences that make up more than 0.1% of the total number of sequences). | The file is marked as not meeting the metric criteria. |
| Adapter contaminated | The file contains no more than 1% reads contaminated with adapter sequences. | The "Quality Trimmer" task: removing adapters. |
| Base N content | Among all nucleotides in the file, there are no more than 20% unknown (N) nucleotides. | The file is marked as not meeting the metric criteria. |
| GC content | The threshold for detecting GC-content disturbances in the file is 0.035. The acceptable length of GC-content disturbances in the file is 4. | The file is marked as not meeting the metric criteria. |
| Base sequence content | The threshold for the maximum difference from all reads maximum between A and T, or G and C in the file is 20%. The threshold for the difference and the mean of difference between A and T, or G and C in the file is 1%. | The file is marked as not meeting the metric criteria. |
Cleanup if enabled:
2.1. Filter by Tile if there are more than 10 faulty tiles (i.e. tiles that exceed mean quality across all tiles by 7). During filtering, tiles that contain reads that exceed the maximum mean quality are excluded from the analysis using BBMap FilterByTile. After filtering by Tile, the Check Quality is performed again.
2.2. Quality Trimmer:
- Remove adapters if the sample file contains more than 1% of reads contaminated with adapter sequences.
- Filter short reads if the sample file contains more than 25% of short reads (i.e. reads shorter than the short sequence bond (20 bp)).
- Trim by quality if the quality of the three end positions in a read is below the minimal base percentile quality (20).
Quality Trimmer is performed using Cutadapt. After quality trimming, the Check Quality is performed again, and if the sample still does not meet the quality criteria, the "Quality Trimmer - Check Quality" cycle is repeated iteratively with different parameter values (quality threshold and minimum length) until the sample meets the quality criteria.
2.3. Build Sample: if a sample requires quality trimming but contains more than 600,000 reads, then a test sample is first constructed, consisting of each n-th read of the sample, where n is the sampling frequency (specified in the task parameters). You can download the built sample file in the "Result files" section in the "Build Sample" task details ("Download FASTQ_GZ"). The built sample undergoes Check Quality, and then the "Quality Trimmer - Check Quality" cycle is iteratively repeated for it with different parameter values (quality threshold and minimum length) until the sample meets the quality criteria. After that, Quality Trimmer and subsequent Check Quality are launched for the original sample file with the parameters that were calculated for the test sample.
2.4. Quality Filter: if the quality of the three start positions in a read is below the minimal base percentile quality (20), low-quality reads are filtered using FASTX-Toolkit Quality Filter. After filtering, the Check Quality is performed again, and if the sample still does not meet the quality criteria, the "Quality Filter - Check Quality" cycle is repeated iteratively with different parameter values (minimum quality score to keep and minimum percent of bases) until the sample meets the quality criteria.
Tasks from iterative quality trimming and filtering cycles whose parameters were not suitable for obtaining
results that meet the quality criteria, as well as the "Build Sample" task, have the
status , since they are not directly
related to sample analysis.
You can download a sample file after cleanup in the "Result files" section in the details of the cleanup task ("Filter by Tile", "Quality Trimmer", "Quality Filter") that was completed last ("Download FASTQ_GZ").
If alignment is included in the sample analysis workflow, the "Alignment" stage will begin after the "Check quality and cleanup" stage has been successfully completed.