Copy number variations discovery
After the "Variant discovery preprocessing" stage has been successfully completed, copy number variations (CNVs) discovery, which determines how many copies of a DNA region were, on average, present in the sequenced genome, can be initiated for a sample. To run the stage, it must be included in the analysis workflow and the sequencing type and capture kit must be defined for the sample (either automatically or manually by the user). The copy number variations discovery pipeline differs for non-tumor or single tumor samples, and tumor sample from a tumor/control sample set.
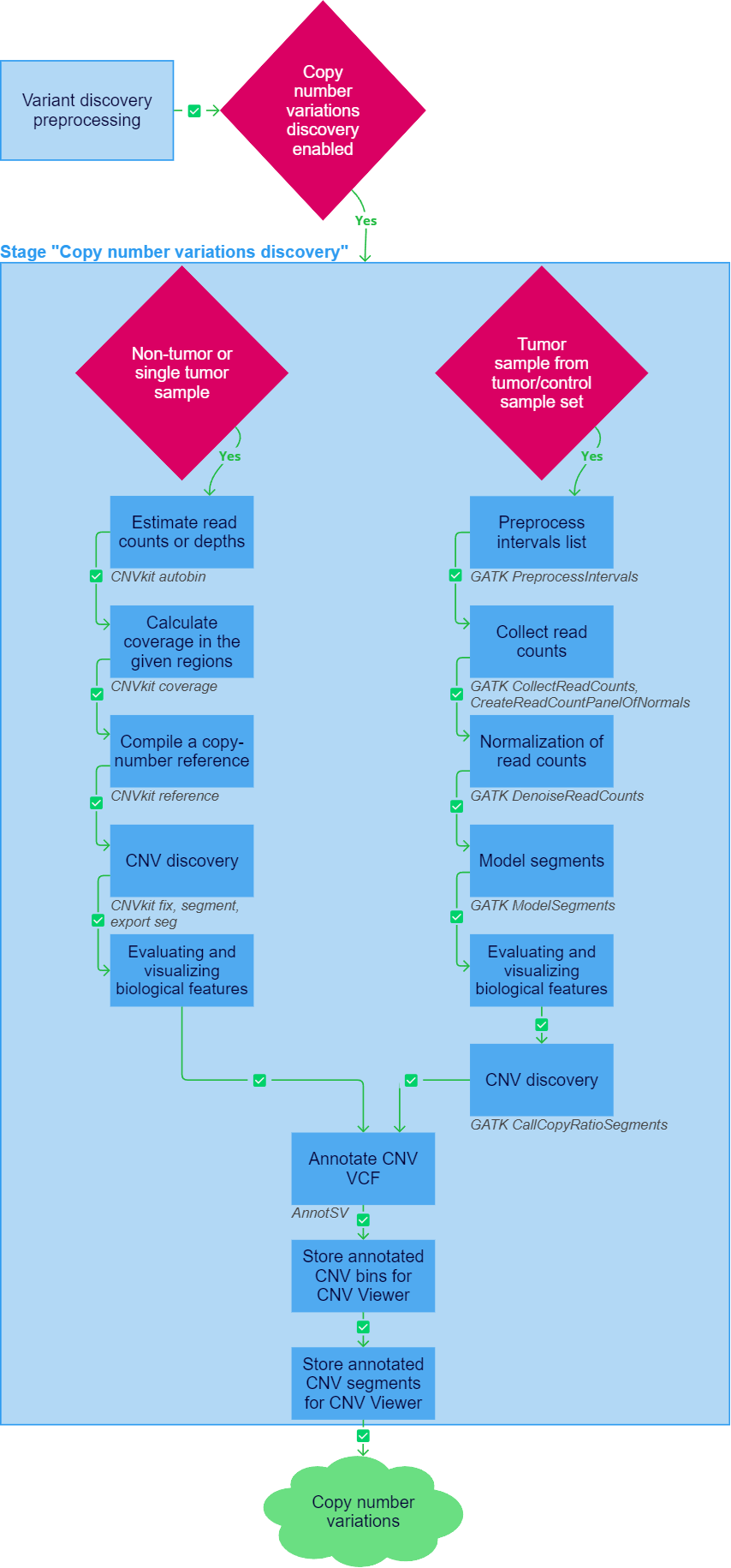
Non-tumor or single tumor sample#
In case of analysis of a tumor sequencing sample only without control or prenatal diagnosis (analysis of a non-tumor sequencing sample), CNVkit tools are used to discover copy number variations.
1. Estimate read counts or depths#
In the first step, CNVkit autobin estimates read counts or depths in an alignment file to estimate reasonable on- and off-target bin (i.e. a coverage assessment interval) sizes. In addition, it adds gene names to target regions. Depending on the coverage depth and the sequencing type, different ways of dividing the genome into bins, within which reads will be counted, are applied:
- If the sample sequencing type is defined as "Panel" (sequencing using a targeted panel) or "WES" (whole-exome sequencing), autobin is provided with the uniquely mapped genome regions and the capture kit intervals corresponding to the sample (i.e. the targeted panel used for targeted selection).
- If the sample sequencing type is defined as "WGS" (whole-genome sequencing) or "Low-pass WGS" (whole genome sequencing with low coverage), autobin is provided with the genomic regions available for sequencing. First, a target bin size is calculated in the range from 10 to 1000000000 bp. Then, the estimated bin size is used to limit the given bin size, which is defined in parameters. This size is used for generating an output.
As a result, autobin generates target and antitarget BED files, and prints a table of estimated average read depths and recommended bin sizes.
If a reference samples panel for WGS or for Targeted capture is selected for sample analysis, the bin size corresponding to the selected panel (it can be customized for some panels) will be used instead of estimating target bin sizes.
The resulting files with target and antitarget bins can be downloaded in the "Result files" section in the "Estimate read counts or depths" task details ("Download target BED" and "Download antitarget BED", respectively).
info
If the sample analysis reveals that the sequencing sample has too low coverage with the capture kit (more than 10% of capture kit regions have zero coverage), then the copy number variations discovery is not further performed for the sample. This can happen if the capture kit was incorrectly selected or if the sample is of low quality.
2. Calculate coverage in the given regions#
At this step, CNVkit coverage calculates the number of reads in each of the bins determined in the previous step. Coverage is calculated via mean read depth from a pileup. The resulting files with coverage information in target and antitarget bins can be downloaded in the "Result files" section in the "Calculate coverage in the given regions" task details ("Download target coverage CNN" and "Download antitarget coverage CNN", respectively).
3. Compile a copy-number reference#
To discover copy number variations by read depth, it is necessary to estimate the depth deviation from the copy-number reference. Normal coverage in regions is calculated taking into account the GC content and repeat-masked proportion of each region. This is done using the CNVkit reference tool. Both target and antitarget bins are involved in the compilation.
If a reference samples panel for WGS or for Targeted capture is selected for sample analysis, then this panel will be used instead of compiling a copy-number reference.
The resulting file with the expected read depth and the reliability of this estimate can be downloaded in the "Result files" section in the "Compile a copy-number reference" task details ("Download CNN").
4. CNV discovery#
This is the key step where the copy number of each genome region is calculated based on the number of reads mapped to the genome. For this, CNVkit tools are used:
- CNVkit fix combines the uncorrected target and antitarget coverage tables and correct for biases in regional coverage and GC content, according to the reference. The resulting table of copy number ratios can be downloaded in the "Result files" section in the "CNV discovery" task details ("Download CNR").
- CNVkit segment infers discrete copy number segments from the coverage table. If the sample sequencing type is a targeted panel, or if a single tumor sample is analyzed, then the circular binary segmentation (CBS) is used as the segmentation algorithm. If the sample sequencing type is WGS, then the experimental method is used as the segmentation algorithm, that is a 3-state hidden Markov model with fixed amplitude for the loss, neutral, and gain states corresponding to absolute copy numbers of 1, 2, and 3. The resulting file with segments can be downloaded in the same section ("Download CNS").
- CNVkit export seg exports the segmentation files to the standard SEG format to be loaded in the Integrative Genomics Viewer (IGV). Called segments in SEG format can be downloaded in the same section ("Download Called segments SEG").
5. Evaluating and visualizing biological features#
At this step, the copy number variations are processed depending on the log2FC threshold value, that is the logarithmic ratio of the detected copy number to the normal copy number. The default threshold value (-0.7) can be changed in the parameters. Using this value as an example, let's see how the thresholds for calling deletions and duplications on autosomes and sex chromosomes will be calculated:
- Copy number variations on autosomes with log2FC ≤ -0.7 are considered deletions. -0.7 is the default log2FC threshold, below which only one autosome is called instead of the normal two.
- Copy number variations on the X and Y chromosomes with log2FC ≤ -2.11 are considered deletions.
- Copy number variations on autosomes with log2FC ≥ 0.46 are considered duplications. The threshold for duplication on an autosome is calculated using the following formula: log2FC = log2((2+D)/2), where D is the “number” of copy number that is “lost” at log2FC = -0.7, and 2 is the number of sister chromatids in the autosome.
- Copy number variations on the X and Y chromosomes with log2FC ≥ 0.82 are considered duplications.
- Variations that do not satisfy the thresholds, i.e. variations on autosomes with -0.7 < log2FC < 0.46 and variations on sex chromosomes with -2.11 < log2FC < 0.82 do not pass filtering.
Click here to see how log2FC thresholds are calculated for deletions and duplications on autosomes and sex chromosomes
Delta of copy number: D = 2 – C = 2 – 2 × 2T21.
Then the corresponding threshold of calling three copies instead of two on autosomes is calculated as follows: T23 = log2((2+D)/2) = log2(2-2T21).
Threshold of calling deletions on autosomes: lo = T21.
Threshold of calling duplications on autosomes: hi = T23.
To calculate the threshold of calling three copies instead of two on the X chromosome, we normalize to XY genotype by dividing it by copy number of 1: T23X= log2((2+D)/1) = log2(2+2-2×2T21) = 1 + log2(2-2T21) = 1 + T23.
Calculating the threshold of calling duplications on the X chromosome:
- if there is no Y chromosome: Xhi = 1 + T23;
- if there is Y chromosome: Xhi = log2(3-2×2T21).
The threshold of calling one copy instead of two on the X chromosome: T21X = log2((2-D)/1) = log2(2-2+2×2T21) = 1 + T21.
Threshold of calling no copies instead of one on the X chromosome: T10X = log2((1-D)/1) = log2(1-2+2×2T21) = log2(-1+2×2T21).
Calculating the threshold of calling deletions on the X chromosome:
- if there is no Y chromosome: Xlo = 1 + T21;
- if there is Y chromosome: Xlo = log2(-1+2×2T21).
The threshold of calling one copy instead of none on the Y chromosome in the case of genotype XX: T01 = log2((0+D)/1) = log2(2-2×2T21) = log2(2×(1-2T21)) = 1 + log2(1-2T21).
Calculating the threshold of calling duplication Yhi and deletions Ylo on the Y chromosome:
- if there is no Y chromosome: Yhi = 1 + log2(1-2T21)); Ylo = -∞;
- if there is Y chromosome: Yhi = Xhi; Ylo = Xlo.
If the CNV aneuploidies calling only is enabled for a non-tumor sample, then only aneuploidies (copy number variations on chromosomes) will be presented in the resulting files.
As a result of processing the variations, the following files are created, which can be downloaded in the "Results files" section in the "Evaluating and visualizing biological features" task details:
- The CNV coverage file in TSV format, that is an unannotated file convenient for studying copy number variations of large regions (chromosome arms, chromosomes). File download link: "Download CNV coverage TSV". You can also open this file in Google Sheets.
- A file with copy number bins in BED format ("Download CNV bins BED").
- A file with copy number segments in BED format ("Download CNV segments BED").
- A file with log2FC thresholds, i.e. the logarithmic ratio of the detected copy number to the normal copy number, based on which copy number variations are filtered. The log2FC values at which variations on autosomes, the X or Y chromosome are considered deletions or duplications are provided, too. File download link: "Download CNV thresholds JSON".
In addition, files with genome-wide visualization and karyogram are generated, which are convenient for studying large copy number variations. You can open them on the "Main" tab or download in the "Results files" section in the "Evaluating and visualizing biological features" task details:
- Karyogram-like graph with denoted chromosome-level CNVs in JPG format ("Download Karyogram-like graph JPG").
- Genome-wide CNV segments' plot in JPG format ("Download JPG").
- PDF file with CNV segment graphs for all chromosomes ("Download PDF").
- Files with CNV segment graphs for each chromosome separately ("Download [chromosome] JPG").
6. Annotate CNV VCF#
To completely and effectively interpret the CNV impact on the phenotype, each detected variation is annotated. Annotation and ranking of discovered copy number variations is performed using the AnnotSV program. This tool compiles functionally, regulatory and clinically relevant information and aims at providing annotations useful to interpret CNV potential pathogenicity and filter out CNV potential false positives. Segments and bins are annotated separately.
The resulting files with the annotated copy number variations can be downloaded in the "Result files" section in the "Annotate CNV VCF" task details:
- A file with all copy number segments in TSV format ("Download Segments all TSV"). You can also open this file in Google Sheets.
- A file with all copy number segments in CSV format ("Download Segments all CSV").
- A file with filtered copy number segments in TSV format ("Download Segments filtered TSV"). You can also open this file in Google Sheets.
- A file with filtered copy number segments in CSV format ("Download Segments filtered CSV").
- A file with all copy number bins in TSV format ("Download Bins all TSV"). You can also open this file in Google Sheets.
- A file with all copy number bins in CSV format ("Download Bins all CSV").
- A file with filtered copy number bins in TSV format ("Download Bins filtered TSV"). You can also open this file in Google Sheets.
- A file with filtered copy number bins in CSV format ("Download Bins filtered CSV").
You can find the description of annotation format and fields in the AnnotSV documentation.
7. Store annotated CNV bins for CNV Viewer#
Saving the results of copy number variations discovery in bins for display in the embedded service for viewing and analyzing copy number variations CNV Viewer.
8. Store annotated CNV segments for CNV Viewer#
Saving the results of copy number variations discovery in segments for display in the embedded service for viewing and analyzing copy number variations CNV Viewer.
After the "Copy number variations discovery" stage, the analysis can proceed with report generation.
Tumor sample from a tumor/control sample set#
In case of analyzing a tumor sequencing sample from a tumor and matched normal sample pair, the GATK pipeline is used to to discover copy number variations.
1. Preprocess intervals list#
GATK PreprocessIntervals prepares bins for coverage collection.
The way of bin preparation varies depending on the sample sequencing type:
- If the sample sequencing type is defined as "Panel" (sequencing using a targeted panel) or "WES" (whole-exome sequencing), the input intervals (capture kit) are first checked for overlapping intervals, which are merged. Then the resulting intervals are padded and binned. Bins containing only N (unknown nucleotides) are filtered out. Binning (grouping intervals into a smaller number of bins) is not performed. Abutting intervals (i.e. intervals that are directly side-by-side but do not actually overlap) are not merged, but treated as separate intervals.
- If the sample sequencing type is defined as "WGS" (whole-genome sequencing) or "Low-pass WGS" (whole genome sequencing with low coverage), each contig is taken as one interval and grouped accordingly. Abutting intervals are merged into a single continuous interval. This creates bins suitable for whole genome sequencing analysis. The length of the generated bins is controlled by the corresponding parameter. Bins containing only N (unknown nucleotides) are filtered out.
The resulting preprocessed Picard interval-list file can be downloaded in the "Result files" section in the "Preprocess intervals list" task details ("Download INTERVAL_LIST").
2. Collect read counts#
GATK CollectReadCounts collects read counts at specified intervals. The count for each interval is calculated by counting the number of read starts that lie in the interval. Abutting intervals (i.e. intervals that are directly side-by-side but do not actually overlap) are not merged, but treated as separate intervals. The command is executed for the alignment files of the tumor and matched normal samples. The resulting counts files in HDF5 format (Hierarchical Data Format, Version 5) can be downloaded in the "Result files" section in the "Collect read counts" task details (the result for the normal sample is "Download Normal file read counts HDF5", the result for the tumor sample is "Download Tumor file read counts HDF5").
GATK CreateReadCountPanelOfNormals creates a panel of normals (PoN) for read-count denoising given the read counts for samples in the panel. It is carried out by counting the number of reads in the normal sample. Genomic intervals with a median (across samples) of fractional coverage less than or equal to 5th percentile are filtered out. The resulting panel-of-normals file can be downloaded in the "Result files" section in the "Collect read counts" task details ("Download Generated panel of normal HDF5").
3. Normalization of read counts#
GATK DenoiseReadCounts denoises read counts to produce denoised copy ratios. A panel of normals produced in the previous step is used to standardize and denoise read counts in the tumor sample. Standardization includes (a) transforming to fractional coverage, (b) filtering intervals to those contained in the panel, (c) dividing by interval medians contained in the panel, (d) dividing by the sample median, and (e) transforming to log2 copy ratio. The result is then denoised by subtracting the projection onto the specified number of principal components from the panel. The output is the following files, which can be downloaded in the "Result files" section in the "Normalization of read counts" task details:
- Denoised-copy-ratios file in TSV format ("Download Denoised copy ratios TSV"). You can also open this file in Google Sheets.
- Standardized-copy-ratios file in TSV format ("Download Standardized copy ratios TSV"). You can open this file in Google Sheets, too.
4. Model segments#
GATK ModelSegments models segmented copy ratios from denoised read counts. Segmentation is done using kernel segmentation. Then Markov-chain Monte Carlo (MCMC) is applied to determine posteriors for segmented models for the log2 copy ratio. Finally, smoothing of the segmented posteriors is performed by merging adjacent segments whose posterior credible intervals sufficiently overlap sufficiently (number of 10% equal-tailed credible-interval widths must be equal to 2). Additional rounds of segmentation smoothing are performed until convergence, at which point a final round of MCMC is performed.
Copy-ratio segment file can be downloaded in the "Result files" section in the "Model segments" task details ("Download Copy ration segments SEG"). A file with modeled segments after segmentation smoothing can be downloaded in the same section ("Download Modeled segments SEG").
5. Evaluating and visualizing biological features#
At this step, the copy number variations are processed depending on the log2FC threshold value, that is the logarithmic ratio of the detected copy number to the normal copy number. The default threshold value (-0.7) can be changed in the parameters. Using this value as an example, let's see how the thresholds for calling deletions and duplications on autosomes and sex chromosomes will be calculated:
- Copy number variations on autosomes with log2FC ≤ -0.7 are considered deletions. -0.7 is the default log2FC threshold, below which only one autosome is called instead of the normal two.
- Copy number variations on the X and Y chromosomes with log2FC ≤ -2.11 are considered deletions.
- Copy number variations on autosomes with log2FC ≥ 0.46 are considered duplications. The threshold for duplication on an autosome is calculated using the following formula: log2FC = log2((2+D)/2), where D is the “number” of copy number that is “lost” at log2FC = -0.7, and 2 is the number of sister chromatids in the autosome.
- Copy number variations on the X and Y chromosomes with log2FC ≥ 0.82 are considered duplications.
- Variations that do not satisfy the thresholds, i.e. variations on autosomes with -0.7 < log2FC < 0.46 and variations on sex chromosomes with -2.11 < log2FC < 0.82 do not pass filtering.
Click here to see how log2FC thresholds are calculated for deletions and duplications on autosomes and sex chromosomes
Delta of copy number: D = 2 – C = 2 – 2 × 2T21.
Then the corresponding threshold of calling three copies instead of two on autosomes is calculated as follows: T23 = log2((2+D)/2) = log2(2-2T21).
Threshold of calling deletions on autosomes: lo = T21.
Threshold of calling duplications on autosomes: hi = T23.
To calculate the threshold of calling three copies instead of two on the X chromosome, we normalize to XY genotype by dividing it by copy number of 1: T23X= log2((2+D)/1) = log2(2+2-2×2T21) = 1 + log2(2-2T21) = 1 + T23.
Calculating the threshold of calling duplications on the X chromosome:
- if there is no Y chromosome: Xhi = 1 + T23;
- if there is Y chromosome: Xhi = log2(3-2×2T21).
The threshold of calling one copy instead of two on the X chromosome: T21X = log2((2-D)/1) = log2(2-2+2×2T21) = 1 + T21.
Threshold of calling no copies instead of one on the X chromosome: T10X = log2((1-D)/1) = log2(1-2+2×2T21) = log2(-1+2×2T21).
Calculating the threshold of calling deletions on the X chromosome:
- if there is no Y chromosome: Xlo = 1 + T21;
- if there is Y chromosome: Xlo = log2(-1+2×2T21).
The threshold of calling one copy instead of none on the Y chromosome in the case of genotype XX: T01 = log2((0+D)/1) = log2(2-2×2T21) = log2(2×(1-2T21)) = 1 + log2(1-2T21).
Calculating the threshold of calling duplication Yhi and deletions Ylo on the Y chromosome:
- if there is no Y chromosome: Yhi = 1 + log2(1-2T21)); Ylo = -∞;
- if there is Y chromosome: Yhi = Xhi; Ylo = Xlo.
As a result of processing the variations, the following files are created, which can be downloaded in the "Results files" section in the "Evaluating and visualizing biological features" task details:
- The CNV coverage file in TSV format, that is an unannotated file convenient for studying copy number variations of large regions (chromosome arms, chromosomes). File download link: "Download CNV coverage TSV". You can also open this file in Google Sheets.
- A file with copy number bins in BED format ("Download CNV bins BED").
- A file with copy number segments in BED format ("Download CNV segments BED").
- A file with log2FC thresholds, i.e. the logarithmic ratio of the detected copy number to the normal copy number, based on which copy number variations are filtered. The log2FC values at which variations on autosomes, the X or Y chromosome are considered deletions or duplications are provided, too. File download link: "Download CNV thresholds JSON".
In addition, files with genome-wide visualization and karyogram are generated, which are convenient for studying large copy number variations. You can open them on the "Main" tab or download in the "Results files" section in the "Evaluating and visualizing biological features" task details:
- Karyogram-like graph with denoted chromosome-level CNVs in JPG format ("Download Karyogram-like graph JPG").
- Genome-wide CNV segments' plot in JPG format ("Download JPG").
- PDF file with CNV segment graphs for all chromosomes ("Download PDF").
- Files with CNV segment graphs for each chromosome separately ("Download [chromosome] JPG").
6. CNV discovery#
This is the key step where the copy number of each genome region is calculated based on the number of reads mapped to the genome. In case of analyzing a tumor sequencing sample from a tumor and matched normal sample pair, the GATK pipeline is used.
Copy-ratio segments are called as amplified, deleted, or copy-number neutral using GATK CallCopyRatioSegments. This is a relatively naive caller that takes the segments modeled in the "Model segments" step and performs a simple statistical test on the segmented log2 copy ratios to call amplifications and deletions, given a specified range for determining copy-number neutral segments.
The output is the following files, which can be downloaded in the "Result files" section in the "CNV discovery" task details:
- Called copy-ratio-segments file in TSV format ("Download Called segments SEG").
- CBS-formatted (circular binary segmentation) file with information on segments ("Download Statistics SEG"). It is compatible with the Integrative Genomics Viewer (IGV). You can find a description of the file format and its columns here.
7. Annotate CNV VCF#
To completely and effectively interpret the CNV impact on the phenotype, each detected variation is annotated. Annotation and ranking of discovered copy number variations is performed using the AnnotSV program. This tool compiles functionally, regulatory and clinically relevant information and aims at providing annotations useful to interpret CNV potential pathogenicity and filter out CNV potential false positives. Segments and bins are annotated separately.
The resulting files with the annotated copy number variations can be downloaded in the "Result files" section in the "Annotate CNV VCF" task details:
- A file with all copy number segments in TSV format ("Download Segments all TSV"). You can also open this file in Google Sheets.
- A file with all copy number segments in CSV format ("Download Segments all CSV").
- A file with filtered copy number segments in TSV format ("Download Segments filtered TSV"). You can also open this file in Google Sheets.
- A file with filtered copy number segments in CSV format ("Download Segments filtered CSV").
- A file with all copy number bins in TSV format ("Download Bins all TSV"). You can also open this file in Google Sheets.
- A file with all copy number bins in CSV format ("Download Bins all CSV").
- A file with filtered copy number bins in TSV format ("Download Bins filtered TSV"). You can also open this file in Google Sheets.
- A file with filtered copy number bins in CSV format ("Download Bins filtered CSV").
You can find the description of annotation format and fields in the AnnotSV documentation.
8. Store annotated CNV bins for CNV Viewer#
Saving the results of copy number variations discovery in bins for display in the embedded service for viewing and analyzing copy number variations CNV Viewer.
9. Store annotated CNV segments for CNV Viewer#
Saving the results of copy number variations discovery in segments for display in the embedded service for viewing and analyzing copy number variations CNV Viewer.
After the "Copy number variations discovery" stage, the analysis can proceed with report generation.