Alignment
After the "Check quality and cleanup" stage has been successfully completed, further sample analysis (SNVs/Indels, structural variations, or copy number variations discovery, as well as genomic predictions) requires alignment (mapping) of reads to the reference genome. GRCh38.p14, the most recent version of the human genome is used for alignment. If alignment is excluded from the workflow, the analysis is completed after the "Upload, identify and verify" and "Check quality and cleanup" stages have been successfully completed. If any of the tasks listed below fails, the sample analysis is stopped.
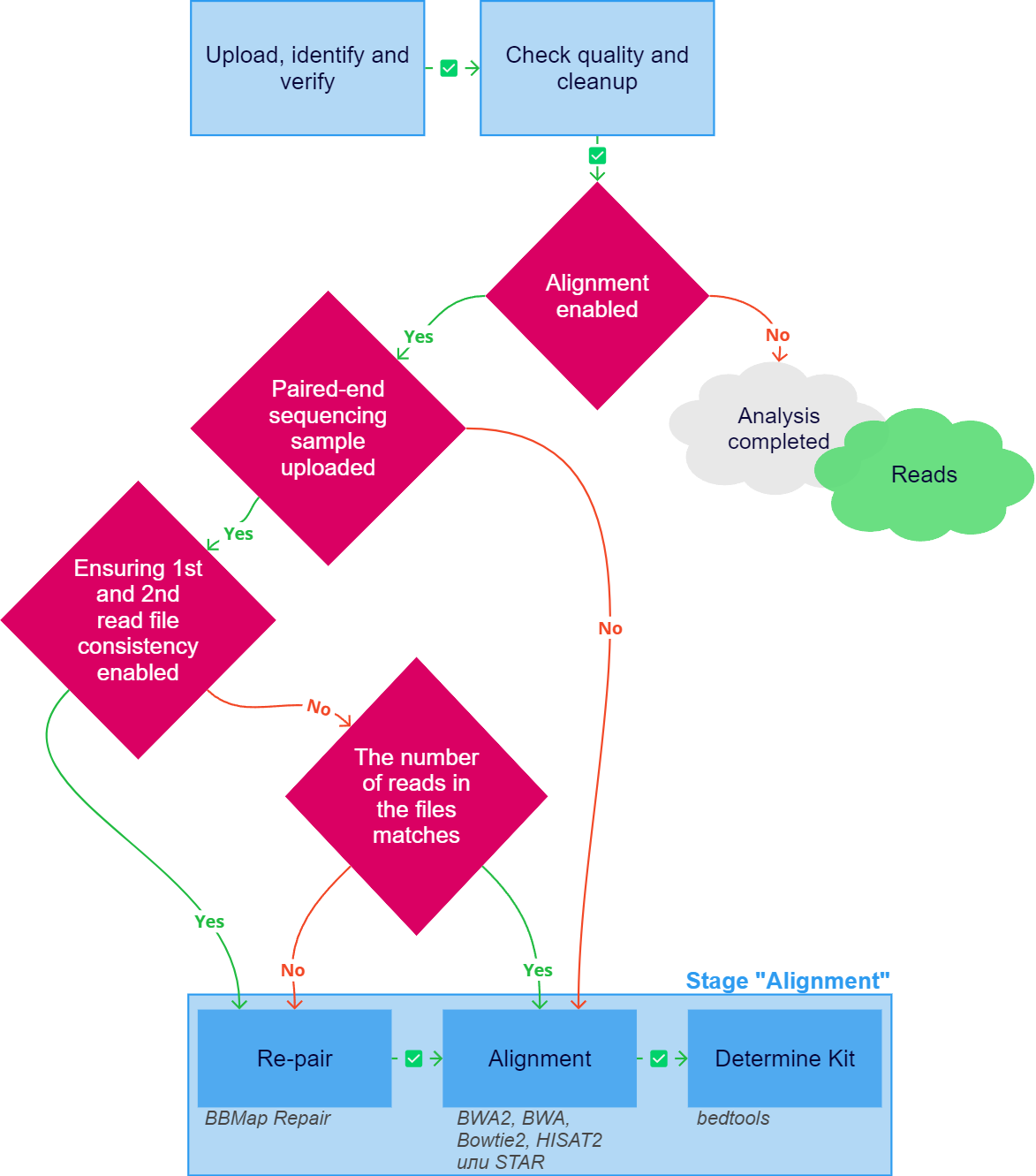
The "Alignment" stage of sample analysis may include the following tasks:
- Re-pair is launched if the following conditions are met:
- A paired-end sequencing sample is being analyzed (i.e. sequencing in which both ends of a DNA fragment are sequenced);
- "Ensure 1st and 2nd read file consistency" parameter is enabled and/or the number of reads in the paired files of the sample does not match.
BBMap Repair is used to re-pair reads. It includes the following:
- Sorting lines in files.
- Restoring the order in paired files: the first read in the first file must match the first read in the second file, etc. The order is restored by analyzing the read names, which must either be in Illumina format (an identical prefix followed by "1:" and "2:" or "/1" and "/2") or completely identical for both reads in a pair.
- Restoring paired reads, which is necessary for alignment, so that each read from one file contains a pair in the other. Paired reads can be broken during cleanup, which leads to the forming of singletons (a singleton is a read with a sequence that is present exactly once, i.e. is unique among the reads) that have lost their pair in the paired file. By default, singletons are removed from the analysis, but they can be used for genome alignment if the corresponding parameter is enabled.
- Alignment is the process of matching sequence reads to a reference sequence (the version hg38 of the reference genome is used). By default, the alignment is performed using the community-recommended tool BWA2 Burrows-Wheeler Aligner (BWA-MEM2), which is the latest and faster version of the BWA-MEM algorithm, which is based on backward search with Burrows–Wheeler Transform (BWT) to efficiently align short sequencing reads against a human genome, allowing mismatches and gaps1. The aligner can be changed in the parameters to BWA Burrows-Wheeler Aligner (i.e. an earlier version of the BWA-MEM2 algorithm2), Bowtie23, HISAT24 or STAR5. The alignment is written to a file in SAM format (i.e. a text-based format for storing biological sequences aligned to a reference sequence).
After alignment, the mapped reads are sorted by leftmost coordinates using samtools sort and written to a file in BAM format (i.e. a binary form of the SAM format, which uses less disk space and is faster to process than SAM). You can download the resulting file in the "Result files" section in the "Alignment" task details ("Download BAM"). You can also open this file in the IGV browser by clicking on the "Open in IGV Browser" link. The BAM file is indexed using samtools index. The resulting index file can be downloaded in the same section ("Download BAI").
info
If you want to add the alignment track obtained from your sample analysis in Genomenal to your desktop IGV, you can do so via a link. To do this, follow these steps:
- Right-click the alignment file link "Download BAM" and select the "Copy link address" option.
- Upload the track via URL to your desktop IGV, as described here.
- Right-click the alignment index file link "Download BAI" and select the "Copy link address" option.
- Add the index via URL to the corresponding field in IGV.
- Click "OK". Done! The sample alignment track is added to IGV.
The "Metrics" section in the "Alignment" task details provides metrics for assessing the quality of alignment:
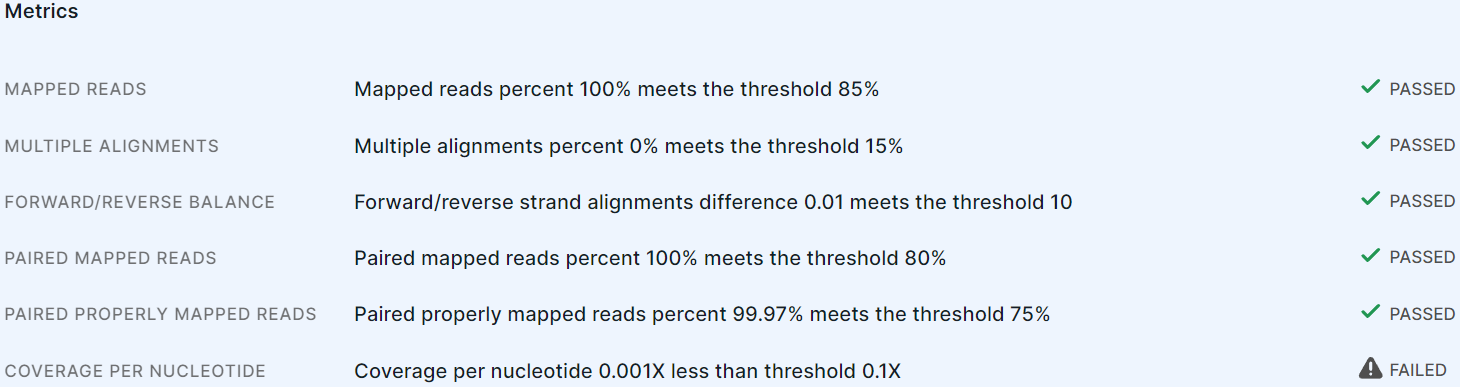
For each metric, the following are provided:
- metric name;
- description of the result of checking the compliance of a certain sample alignment indicator and the threshold of this metric: the indicator value for the sample (e.g. Mapped reads, i.e. the percent of mapped reads in the alignment file) and the used metric threshold, at which the alignment file is considered high-quality (can be changed in the parameters), are provided;
- result of assessing the quality of alignment by
metric:
if the sample alignment satisfies the metric threshold, or
if it does not.
The alignment quality metrics#
| Metric | The metric threshold value at which the alignment file is considered high-quality (this is the default value, can be changed in parameters) |
| Mapped reads | The alignment file contains at least 85% mapped reads. |
| Multiple alignments | The alignment file contains no more than 15% multiple alignments of the same read per genome. |
| Forward/reverse balance | The difference between forward and reverse strand reads is no more than 10% in the alignment file. |
| Paired mapped reads | The alignment file contains at least 80% paired mapped reads. |
| Paired properly mapped reads | The alignment file contains at least 75% paired properly mapped reads. |
| Coverage per nucleotide | The minimum acceptable coverage per nucleotide is 0.1. |
If the metric value does not meet the threshold in the sample alignment file, the file is marked as not meeting the metric criteria, i.e. the alignment is completed successfully, but the stage status indicates the number of metrics that do not meet the criteria. If no sample reads are aligned to the genome, the "Alignment" stage completes with an error and the analysis is stopped.
If at least one of the following stages is included in the sample analysis: somatic SNVs/Indels discovery, germline SNVs/Indels discovery, structural variations discovery, copy number variations discovery, oligogenic risk scores calculation, polygenic risk scores calculation, pharmacogenetic calculation, or ancestry analysis, then after the "Alignment" task is successfully completed, the "Variant discovery preprocessing" stage will start.
- Determine Kit
If the user has defined the sample sequencing type as WGS (whole-genome sequencing), then the capture kit determination is not performed, and the sequencing type can remain WGS or be redefined as Low-pass WGS (whole genome sequencing with low coverage). WGS and Low-pass WGS are differentiated by the average sample coverage: the sequencing type of samples with an average coverage less than 4x is defined as Low-pass WGS, and above or equal to 4x as WGS.
If the user has defined the sample sequencing type as Targeted selection, the sequencing type can be defined as Panel (sequencing using a targeted panel) or WES (whole-exome sequencing). Next, the most suitable capture kit (i.e. the targeted panel used in targeted selection) for the sample is determined or the coverage statistics of the sample by the capture kit selected by the user are calculated:
- bedtools genomecov computes coverage summaries of aligned sequences for a reference genome and reports depth in bedGraph format (File A).
- bedtools intersect screens for overlaps between File A and either the capture kit selected for the sample by the user, or each capture kit that is built into or uploaded into the system if the user has not selected a capture kit. For a capture kit, the coverage depth, kit length, intersection length, complement coverage depth, and complement intersection length of aligned sample sequences with capture kit intervals are calculated.
- For a capture kit selected by the user, or for the best matched capture kit, bedtools complement returns a file with all intervals in a genome that are not covered by at least one interval in the capture kit file (File B).
- bedtools intersect screens for overlaps between File A and File B by computing the coverage index, depth, and intersection length of these files.
As a result, if the user has not selected a capture kit and if the best matched capture kit for the sample was found, the sample is further analyzed with this kit. If a suitable capture kit could not be found, SNVs/Indels filtration (variants discovery in the same genomic intervals in which targeted sequencing was performed) and copy number variation discovery will not be performed for the sample. The file with the statistics of the capture kit selected by the user or determined by the system can be downloaded in the "Result files" section in the "Determine Kit" task details ("Download Capture kit stats JSON").
- Vasimuddin M., Sanchit M., Heng L., Srinivas A. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. IEEE Parallel and Distributed Processing Symposium (IPDPS) (2019)↩
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 (2013)↩
- Langmead B., Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357:359 (2012)↩
- Kim D., Paggi J.M., Park C. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907:915 (2019)↩
- Dobin A., Davis C.A., Schlesinger F. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15:21 (2013)↩