Germline SNVs/Indels discovery and annotation
Germline SNVs/Indels discovery#
After the "Variant discovery preprocessing" stage has been successfully completed, germline SNVs/Indels discovery can be initiated for a non-tumor or single tumor sample if the stage is included in the analysis workflow.
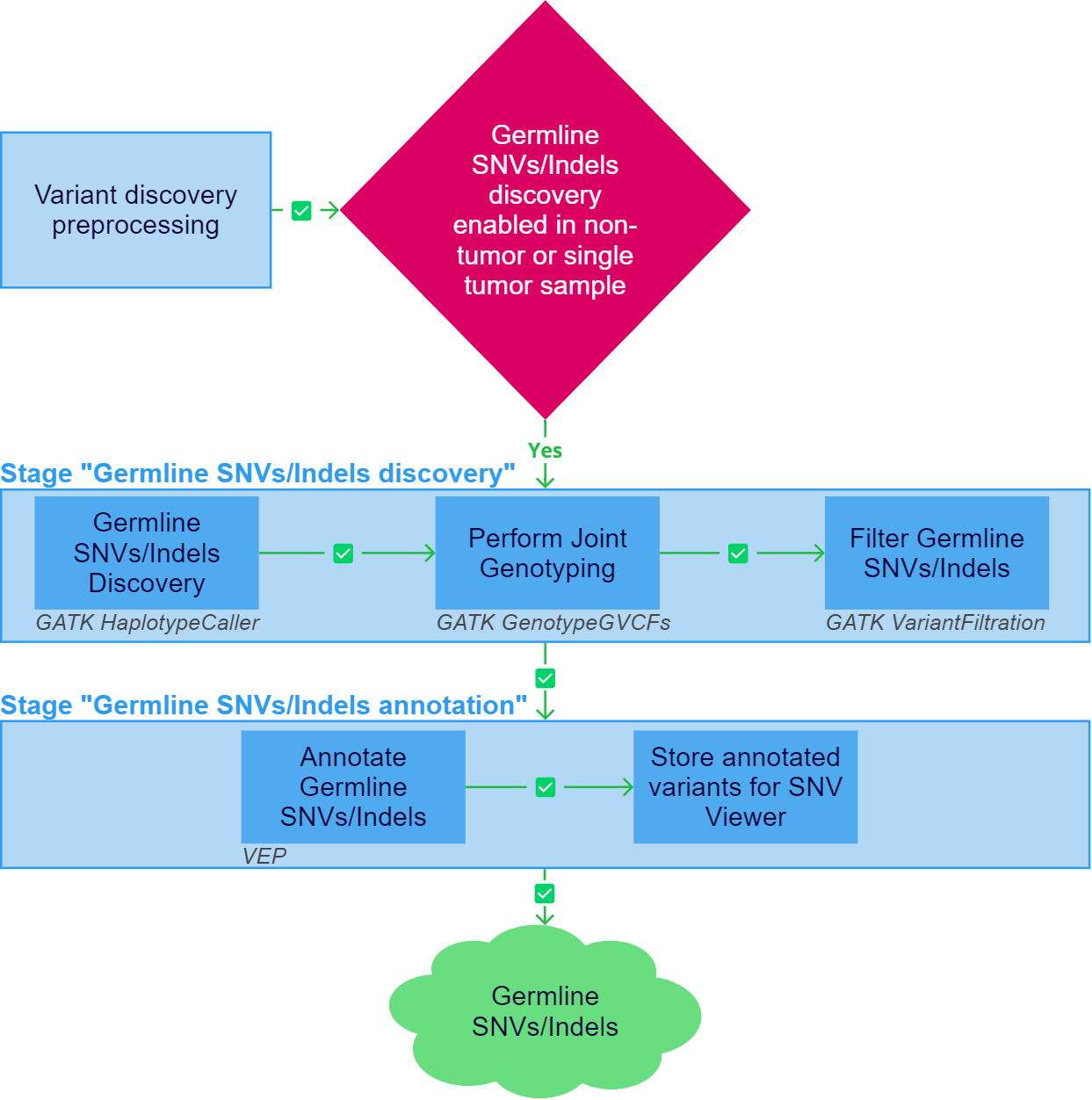
1. Germline SNVs/Indels Discovery#
Germline single-nucleotide variants (SNVs) and short insertions/deletions (indels) calling is performed using GATK HaplotypeCaller.
I. Dividing data into intervals#
To speed up the process and save resources, the Scatter-Gather task divides the input data into 64 genomic intervals. If the sample capture kit is defined, the data is divided using bedtools intersect, which screens for overlaps between distinct genomic intervals and capture kit intervals.
II. Variant calling#
Variant calling in each of the resulting intervals is performed using GATK HaplotypeCaller. Call germline SNPs and indels via local re-assembly of haplotypes Ths tool is capable of calling SNVs and indels simultaneously via local de novo assembly of haplotypes in an active region. In other words, whenever the program encounters a region showing signs of variation, it discards the existing mapping information and completely reassembles the reads in that region. This allows the HaplotypeCaller to be more accurate when calling regions that are traditionally difficult to call, for example when they contain different types of variants close to each other.
Reference model is emitted by homozygous-reference sites compressed into bands of similar genotype quality, i.e. the GVCF (Genomic VCF) format. The key difference between a regular VCF and a GVCF is that the GVCF has records for all sites, whether there is a variant call there or not. The goal is to have every site represented in the file in order to do joint analysis of a cohort in subsequent steps. The records in a GVCF include an accurate estimation of how confident we are in the determination that the sites are homozygous-reference or not.
By default, soft clipped bases (i.e. bases from the ends of reads that do not align with the reference sequence) are analyzed when calling variants. This is controlled by the corresponding setting.
III. Combining GVCF files#
GATK CombineGVCFs combines gVCF files produced by HaplotypeCaller into a single gVCF file. The resulting file is compressed into a GZIP archive using bgzip. It can be downloaded in the "Result files" section in the "Germline SNVs/Indels Discovery" task details ("Download VCF_GZ"). You can also open this file in the IGV browser by clicking on the "Open in IGV Browser" link. In addition, the VCF file is indexed using tabix. The resulting index file can be downloaded in the same section ("Download VCF_TBI").
2. Perform Joint Genotyping#
In the next step, GATK GenotypeGVCFs performs joint genotyping on samples pre-called with HaplotypeCaller. The resulting file is compressed into a GZIP archive using bgzip. It can be downloaded in the "Result files" section in the "Perform Joint Genotyping" task details ("Download VCF_GZ"). You can also open this file in the IGV browser by clicking on the "Open in IGV Browser" link. In addition, the VCF file is indexed using tabix. The resulting index file can be downloaded in the same section ("Download VCF_TBI").
3. Filter Germline SNVs/Indels#
I. Selecting SNVs/Indels#
GATK SelectVariants selects a subset of variants (separately single nucleotide variants and separately indels) from a VCF file. The resulting files with SNVs and with indels are compressed into GZIP archives using bgzip. They can be downloaded in the "Result files" section in the "Filter Germline SNVs/Indels" task details ("Download Unfiltered SNPs VCF_GZ" and "Download Unfiltered INDELs VCF_GZ", respectively). You can also open these files in the IGV browser by clicking on the "Open in IGV Browser" link. In addition, the VCF files are indexed using tabix. The resulting index files can be downloaded in the same section ("Download Unfiltered SNPs VCF_TBI" and "Download Unfiltered INDELs VCF_TBI").
II. Filtering SNVs/Indels#
Then hard-filtering variant calls based on INFO and/or FORMAT annotations is performed using GATK VariantFiltration. Records are hard-filtered by changing the value in the "FILTER" field to something other than "PASS". Filtered records are preserved in the output. Filtering is performed based on hard-filtering parameters specified for the sample in the settings.
The resulting files with filtered SNVs and filtered indels are compressed into GZIP archives using bgzip. They can be downloaded in the "Result files" section in the "Filter Germline SNVs/Indels" task details ("Download Filtered SNPs VCF_GZ" and "Download Filtered INDELs VCF_GZ", respectively). You can also open these files in the IGV browser by clicking on the "Open in IGV Browser" link. In addition, the VCF files are indexed using tabix. The resulting index files can be downloaded in the same section ("Download Filtered SNPs VCF_TBI" and "Download Filtered INDELs VCF_TBI").
III. Merging filtered variants#
GATK MergeVcfs combines VCF files with filtered single nucleotide variants and filtered indels into a single variant file in VCF format. The resulting file with all germline variants discovered in a sample is compressed into a GZIP archive using bgzip. It can be downloaded in the "Result files" section in the "Filter Germline SNVs/Indels" task details ("Download Filtered SNPs/INDELs VCF_GZ"). You can also open this file in the IGV browser by clicking on the "Open in IGV Browser" link. In addition, the VCF file is indexed using tabix. The resulting index file can be downloaded in the same section ("Download Filtered SNPs/INDELs VCF_TBI").
Germline SNVs/Indels annotation#
After the "Germline SNVs/Indels discovery" stage has been successfully completed, germline SNVs/Indels annotation is performed.
1. Annotate Germline SNVs/Indels#
Annotation of SNVs/Indels in the file with data from the RefSeq, 1000 Genomes, dbNSFP, dbSNP, gnomAD 3, gnomAD 4, ClinVar, CADD, SpliceAI, ENIGMA databases.
Determination of the effect of SNVs/Indels on genes, transcripts, and protein sequence, as well as regulatory regions, using Ensembl Variant Effect Predictor (VEP):
PolyPhen predicts possible impact of an amino acid substitution on the structure and function of a protein using straightforward physical and comparative considerations.
A flag indicating if the transcript is the canonical transcript for the gene is added.
Allele number from VCF input is identified.
Affected exon and intron numbering is added.
HGVS nomenclature based on Ensembl stable identifiers is added.
Upstream variant consequences are determined if the upstream (5') distance between a variant and a transcript is more than 2000 bp. Downstream variant consequences are determined if the downstream (3') distance between a variant and a transcript is more than 1000 bp.
The resulting file with raw annotated SNVs/Indels in TSV format can be downloaded in the "Result files" section in the "Annotate Germline SNVs/Indels" task details ("Download Raw annotated TSV"). You can also open it in Google Sheets.
Files with annotated SNVs/Indels (all variants and filtered variants) without duplicates in TSV format can be downloaded in the same section ("Download All variants TSV" and "Download Filtered variants TSV", respectively). You can also open them in Google Sheets. The same files in CSV format can be downloaded from the same place ("Download All variants CSV" and "Download Filtered variants CSV").
Converting TSV results to VCF.
GATK FixVCFHeader replaces or fixes a VCF header.
The file is compressed into a GZIP archive using bgzip. The resulting files can be downloaded in the "Result files" section in the "Annotate Germline SNVs/Indels" task details (file with all variants: "Download All variants VCF_GZ", file with filtered variants: "Download Filtered variants VCF_GZ"). You can also open these files in the IGV browser by clicking on the "Open in IGV Browser" link.
The files are indexed using tabix. The resulting index files can be downloaded in the same section ("Download All variants VCF_TBI" and "Download Filtered variants VCF_TBI").
Calculating variants' statistic.
2. Store annotated variants for SNV Viewer#
- Saving results for display in SNV Viewer, i.e. an embedded service for viewing and analyzing variants.
- Adding information about the variant occurrence in other samples of the user.
After the "Germline SNVs/Indels annotation" stage, the analysis can proceed with report generation.
info
If you want to add a track with germline SNVs/Indels discovered by the analysis of the sample you uploaded to Genomenal to your desktop IGV, you can do so via a link. Open the details of the necessary task ("Germline SNVs/Indels Discovery", "Perform Joint Genotyping", "Filter Germline SNVs/Indels", "Annotate Germline SNVs/Indels") and do the following:
- Right-click the variant file link (depending on the selected task, this may be the link "Download VCF_GZ", "Download Unfiltered SNPs VCF_GZ", "Download Unfiltered INDELs VCF_GZ", "Download Filtered SNPs VCF_GZ", "Download Filtered INDELs VCF_GZ", "Download Filtered SNPs/INDELs VCF_GZ", "Download Filtered SNPs/INDELs VCF_GZ", "Download All variants VCF_GZ" or "Download Filtered variants VCF_GZ") and select the "Copy link address" option.
- Upload the track via URL to your desktop IGV, as described here.
- Right-click on the download link for the index file corresponding to the annotation file ("Download VCF_TBI", "Download Unfiltered SNPs VCF_TBI", "Download Unfiltered INDELs VCF_TBI", "Download Filtered SNPs VCF_TBI", "Download Filtered INDELs VCF_TBI", "Download Filtered SNPs/INDELs VCF_TBI", "Download Filtered SNPs/INDELs VCF_TBI", "Download All variants VCF_TBI" or "Download Filtered variants VCF_TBI") and select the "Copy link address" option.
- Add the index via URL to the corresponding field in IGV.
- Click "OK". Done! The track with germline SNVs/Indels discovered in the sample is added to IGV.